Featured Manuscripts
For a comprehensive list of Monack Lab publications, please visit PubMed
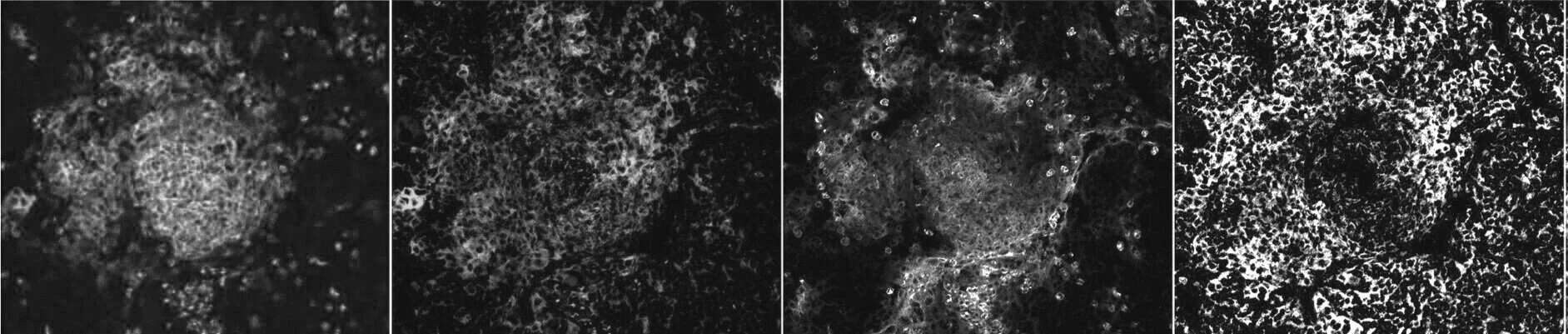
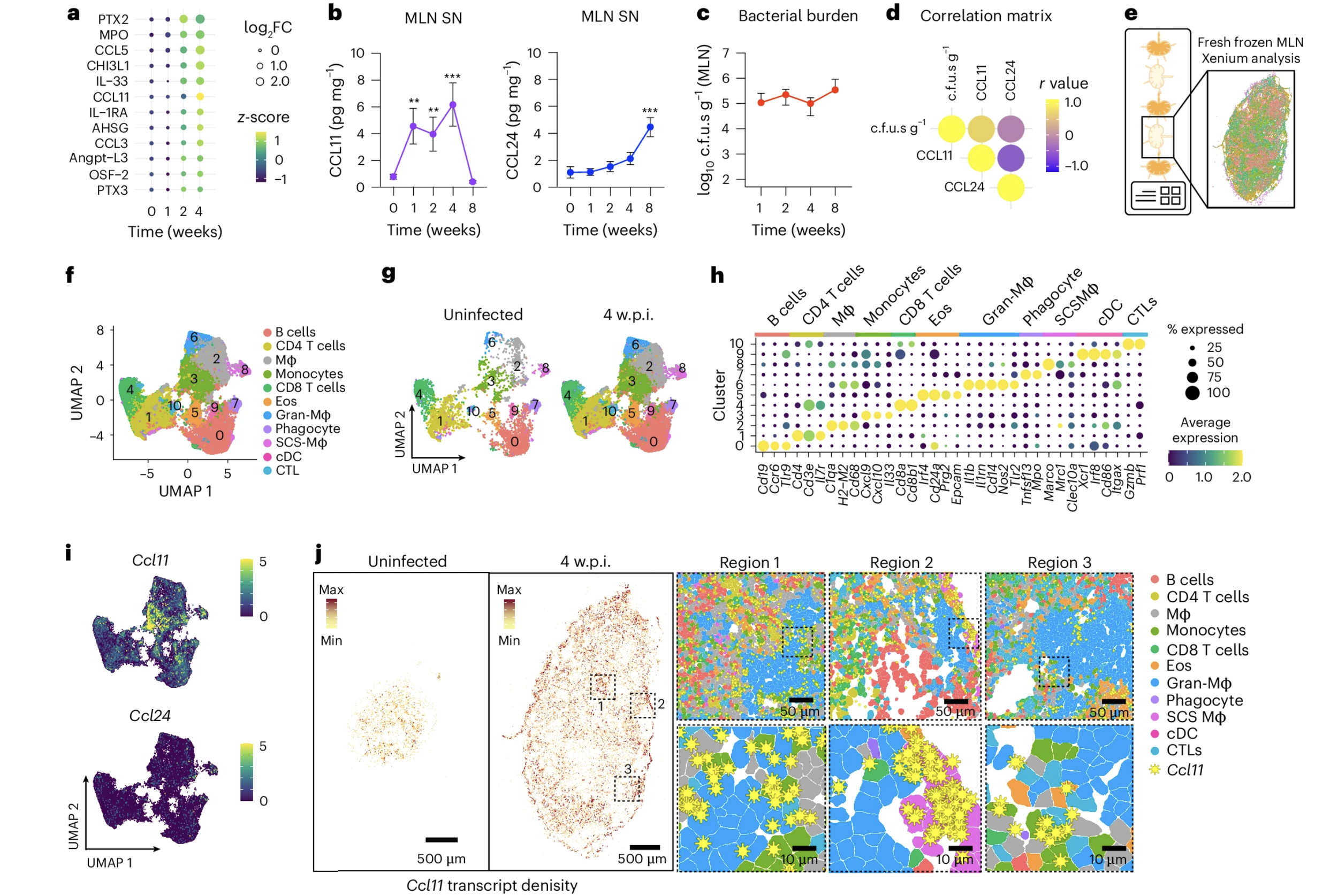
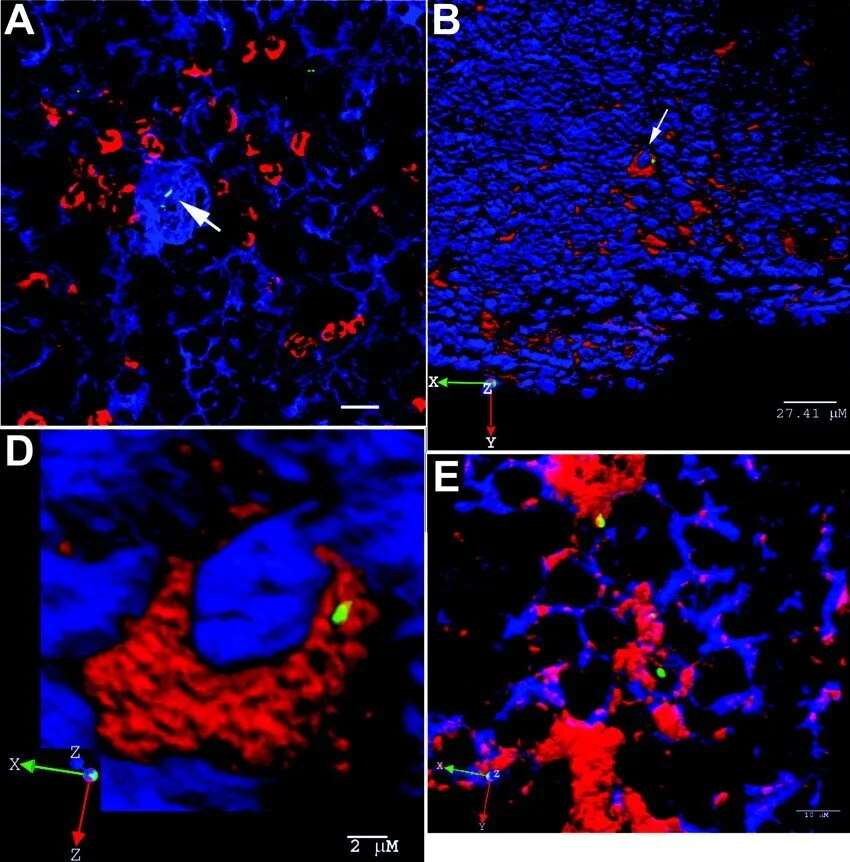
Eosinophils enhance granuloma-mediated control of persistent Salmonella infection in mice
Salmonella enterica can persist asymptomatically within tissues for extended periods. This is achieved through intricate host–pathogen interactions in immune cell aggregates called granulomas, wherein Salmonella establish favourable cellular niches to exploit while the host limits its expansion and tissue dissemination. Here, using a mouse model of persistent Salmonella infection, we identify a host-protective role for eosinophils in the control of Salmonella Typhimurium (STm) infection within the mesenteric lymph nodes, the main lymphoid tissue of STm persistence. Combining spatial transcriptomics and experimental manipulations, we found that monocytes and macrophages responding to STm infection recruited eosinophils in a C-C motif chemokine ligand 11 (CCL11)-dependent manner and enhanced their activation. The protein major basic protein, primarily expressed by eosinophils, was associated with altered macrophage polarization and bacterial control. Thus, eosinophils play a vital role in restraining Salmonella exploitation of granuloma macrophages at a key site of bacterial persistence.
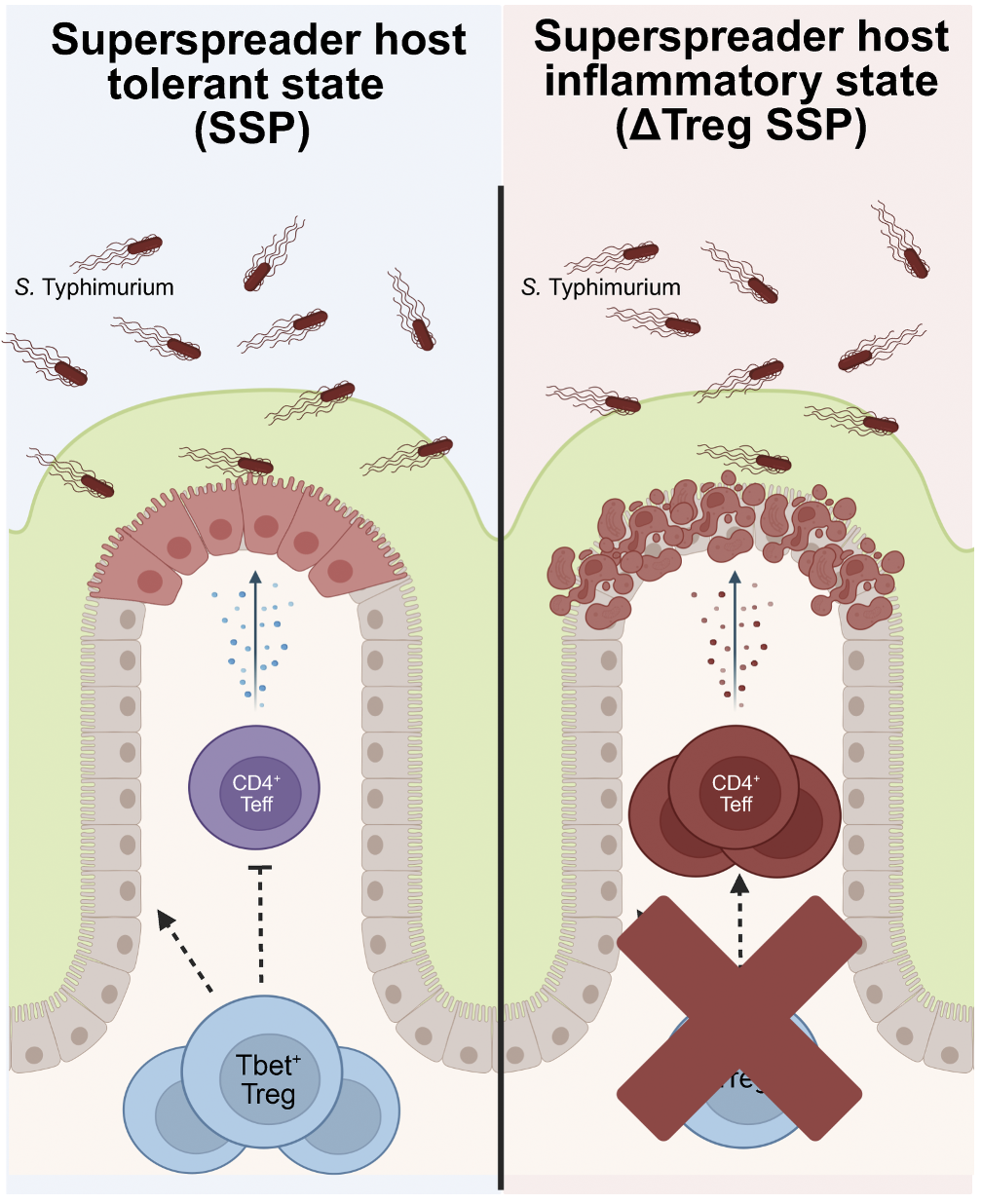
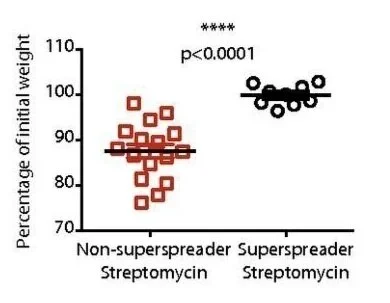
Salmonella-superspreader hosts require gut regulatory T cells to maintain a disease-tolerant state
Journal of Experimental Medicine, 2025
Host–pathogen interactions involve two critical strategies: resistance, whereby hosts clear invading microbes, and tolerance, whereby hosts carry high pathogen burden asymptomatically. Here, we investigate mechanisms by which Salmonella-superspreader (SSP) hosts maintain an asymptomatic state during chronic infection. We found that regulatory T cells (Tregs) are essential for this disease-tolerant state, limiting intestinal immunopathology and enabling SSP hosts to thrive, while facilitating Salmonella transmission. Treg depletion in SSP mice resulted in decreased survival, heightened gut inflammation, and impairment of the intestinal barrier, without affecting Salmonella persistence. Colonic Tregs from SSP mice exhibited a unique transcriptomic profile characterized by the upregulation of type 1 inflammatory genes, including the transcription factor T-bet. In the absence of Tregs, we observed robust expansion of cytotoxic CD4+ T cells, with CD4+ T cell depletion restoring homeostasis. These results uncover a critical host strategy to establish disease tolerance during chronic enteric infection, providing novel insights into mucosal responses to persistent pathogens and chronic intestinal inflammation.
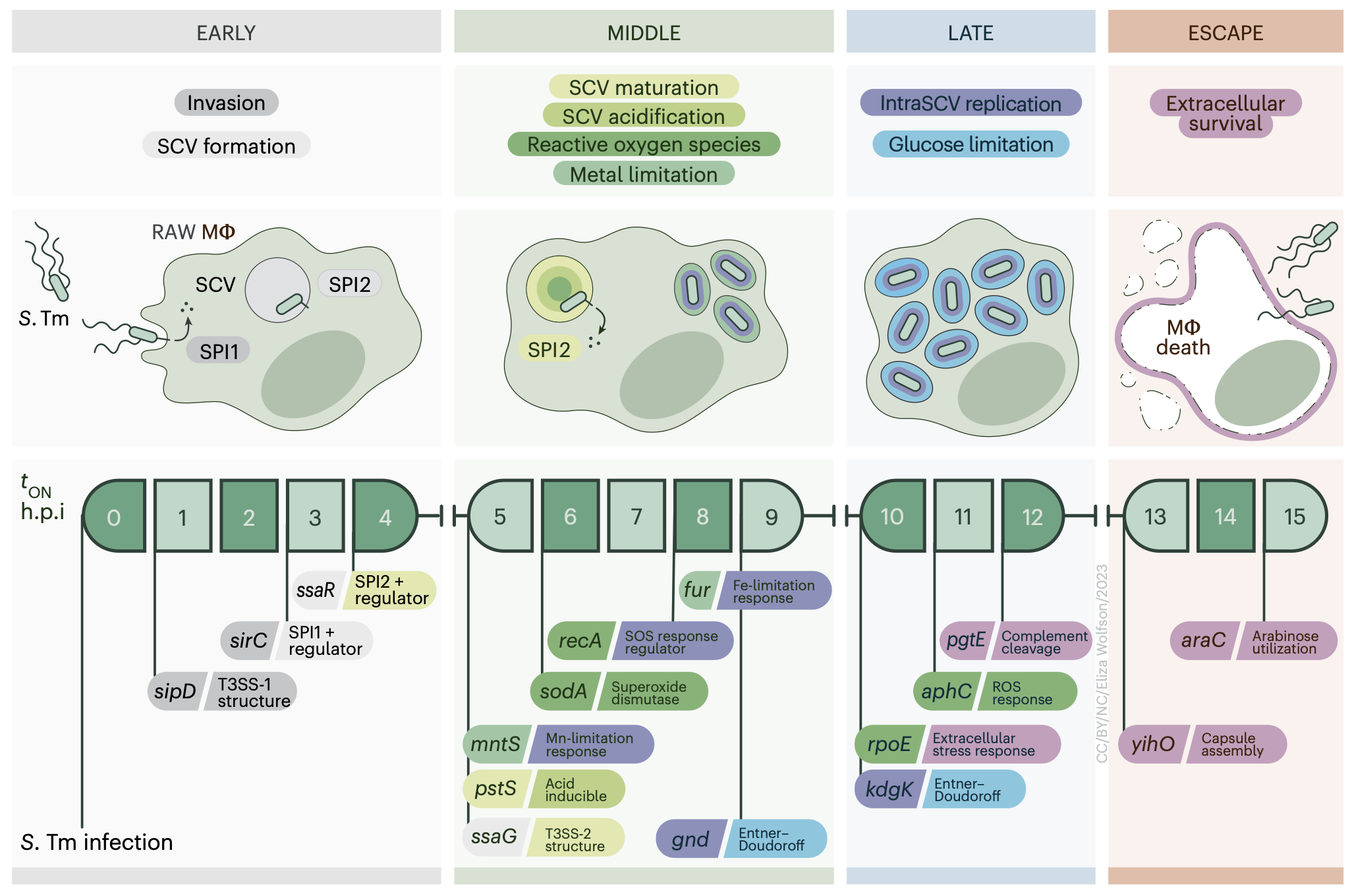
Profiling Salmonella transcriptional dynamics during macrophage infection using a comprehensive reporter library
Salmonella enterica serovar Typhimurium must adapt to rapid environmental shifts, including those encountered upon entry and during replication to survive within macrophages during pathogenesis. Despite extensive RNA-seq-based investigations, questions remain regarding the range, timing and magnitude of response dynamics. Here we constructed a comprehensive GFP-reporter strain library representing 2,901 computationally identified Salmonella promoter regions to study time-resolved Salmonella transcriptional responses. Promoter activity was measured during in vitro growth and during intracellular infection of RAW 264.7 macrophages. Using bulk measurements and single-cell imaging, we uncovered condition-specific transcriptional regulation and population-level heterogeneity in SPI2-related promoter activity. We also discovered previously unidentified transcriptional activity from 234 promoters. These analyses revealed metabolic shifts including requirements for mntS expression to support manganese homeostasis and expression of Entner–Doudoroff pathway-associated genes to support growth within macrophages. Our library and datasets, made available through the online tool SalComKinetics, provide resources for systems-level interrogation of Salmonella transcriptional dynamics.
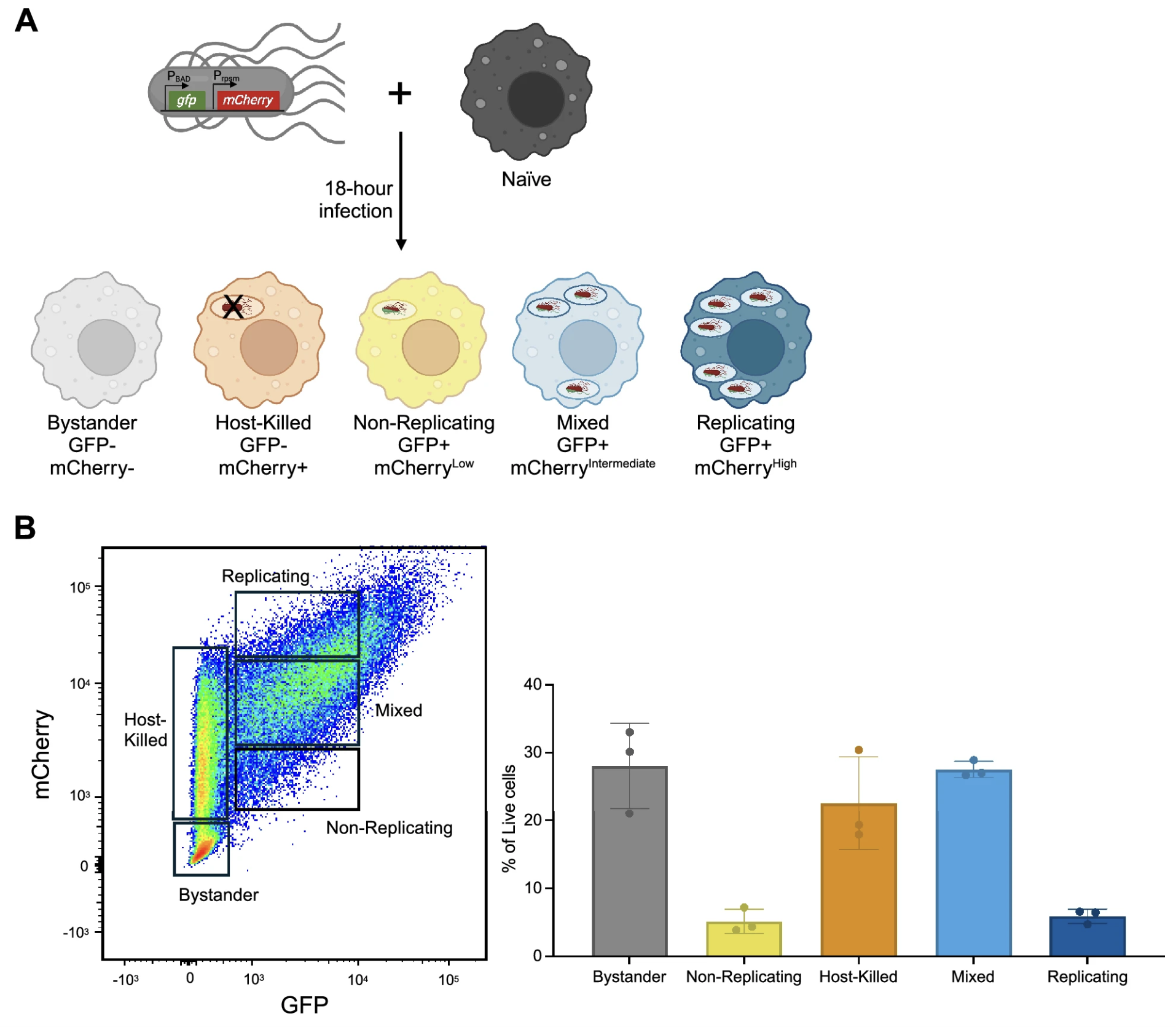
Transcriptional profiling links unique human macrophage phenotypes to the growth of intracellular Salmonella enterica serovar Typhi
Macrophages provide a crucial environment for Salmonella enterica serovar Typhi (S. Typhi) to multiply during typhoid fever, yet our understanding of how human macrophages and S. Typhi interact remains limited. In this study, we delve into the dynamics of S. Typhi replication within human macrophages and the resulting heterogeneous transcriptomic responses of macrophages during infection. Our study reveals key factors that influence macrophage diversity, uncovering distinct immune and metabolic pathways associated with different stages of S. Typhi intracellular replication in macrophages. Of note, we found that macrophages harboring replicating S. Typhi are skewed towards an M1 pro-inflammatory state, whereas macrophages containing non-replicating S. Typhi exhibit neither a distinct M1 pro-inflammatory nor M2 anti-inflammatory state. Additionally, macrophages with replicating S. Typhi were characterized by the increased expression of genes associated with STAT3 phosphorylation and the activation of the STAT3 transcription factor. Our results shed light on transcriptomic pathways involved in the susceptibility of human macrophages to intracellular S. Typhi replication, thereby providing crucial insight into host phenotypes that restrict and support S. Typhi infection.
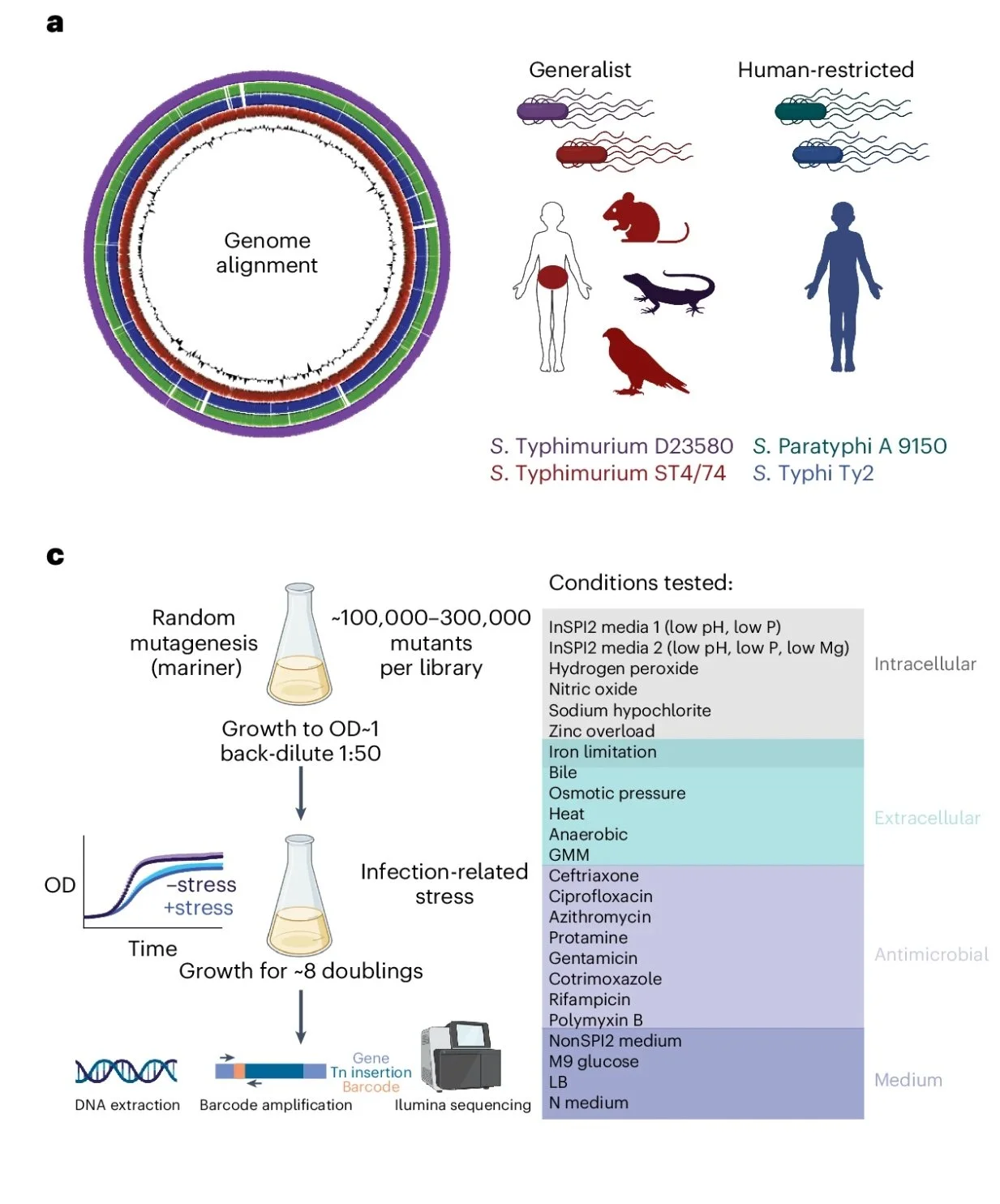
High-throughput fitness experiments reveal specific vulnerabilities of human-adapted Salmonella during stress and infection
Salmonella enterica is comprised of genetically distinct ‘serovars’ that together provide an intriguing model for exploring the genetic basis of pathogen evolution. Although the genomes of numerous Salmonella isolates with broad variations in host range and human disease manifestations have been sequenced, the functional links between genetic and phenotypic differences among these serovars remain poorly understood. Here, we conduct high-throughput functional genomics on both generalist (Typhimurium) and human-restricted (Typhi and Paratyphi A) Salmonella at unprecedented scale in the study of this enteric pathogen. Using a comprehensive systems biology approach, we identify gene networks with serovar-specific fitness effects across 25 host-associated stresses encountered at key stages of human infection. By experimentally perturbing these networks, we characterize previously undescribed pseudogenes in human-adapted Salmonella. Overall, this work highlights specific vulnerabilities encoded within human-restricted Salmonella that are linked to the degradation of their genomes, shedding light into the evolution of this enteric pathogen.
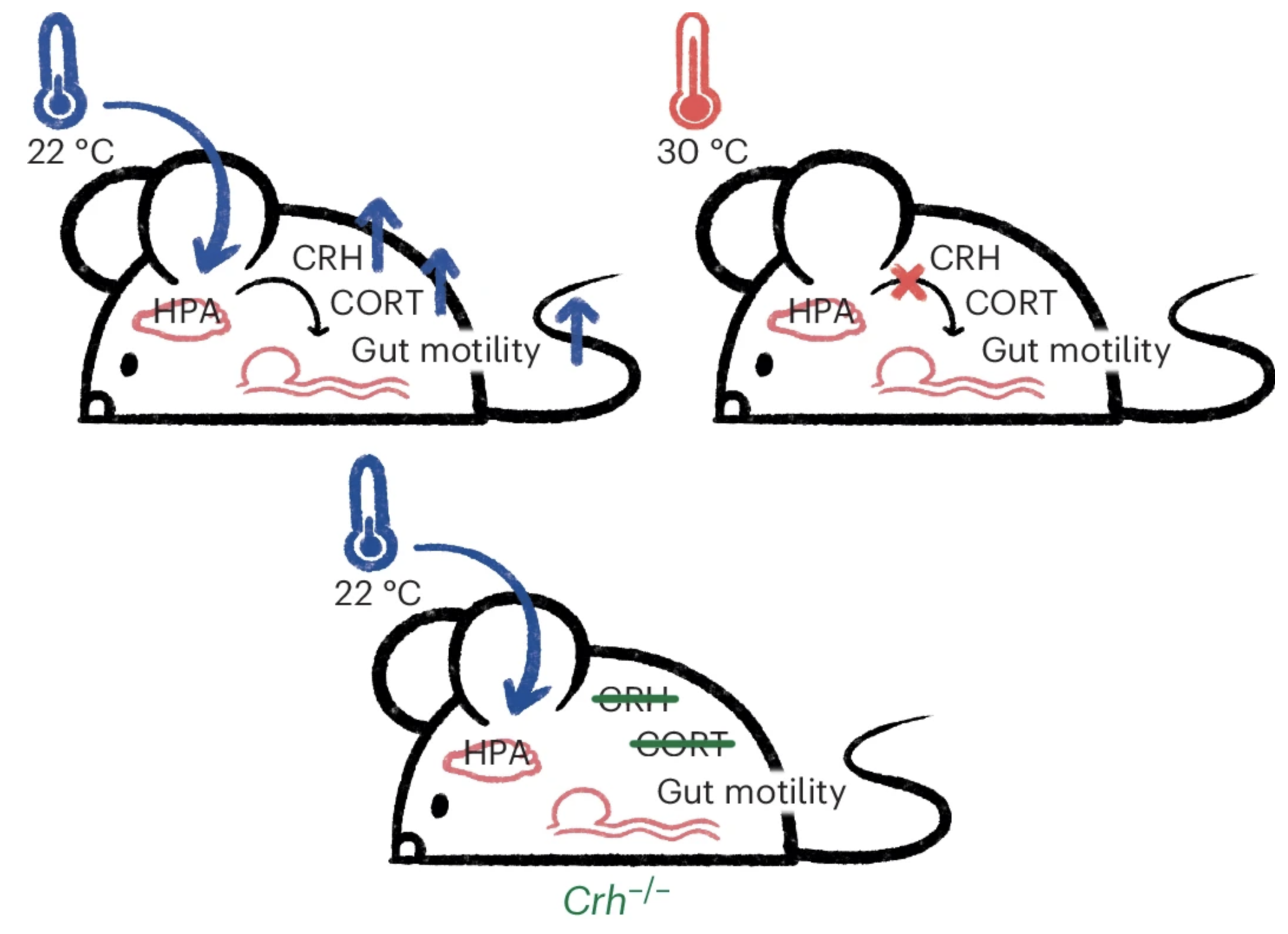
Temperature-dependent differences in mouse gut motility are mediated by stress
Researchers have advocated elevating mouse housing temperatures from the conventional ~22 °C to the mouse thermoneutral point of 30 °C to enhance translational research. However, the impact of environmental temperature on mouse gastrointestinal physiology remains largely unexplored. Here we show that mice raised at 22 °C exhibit whole gut transit speed nearly twice as fast as those raised at 30 °C, primarily driven by a threefold increase in colon transit speed. Furthermore, gut microbiota composition differs between the two temperatures but does not dictate temperature-dependent differences in gut motility. Notably, increased stress signals from the hypothalamic–pituitary–adrenal axis at 22 °C have a pivotal role in mediating temperature-dependent differences in gut motility. Pharmacological and genetic depletion of the stress hormone corticotropin-releasing hormone slows gut motility in stressed 22 °C mice but has no comparable effect in relatively unstressed 30 °C mice. In conclusion, our findings highlight that colder mouse facility temperatures significantly increase gut motility through hormonal stress pathways.
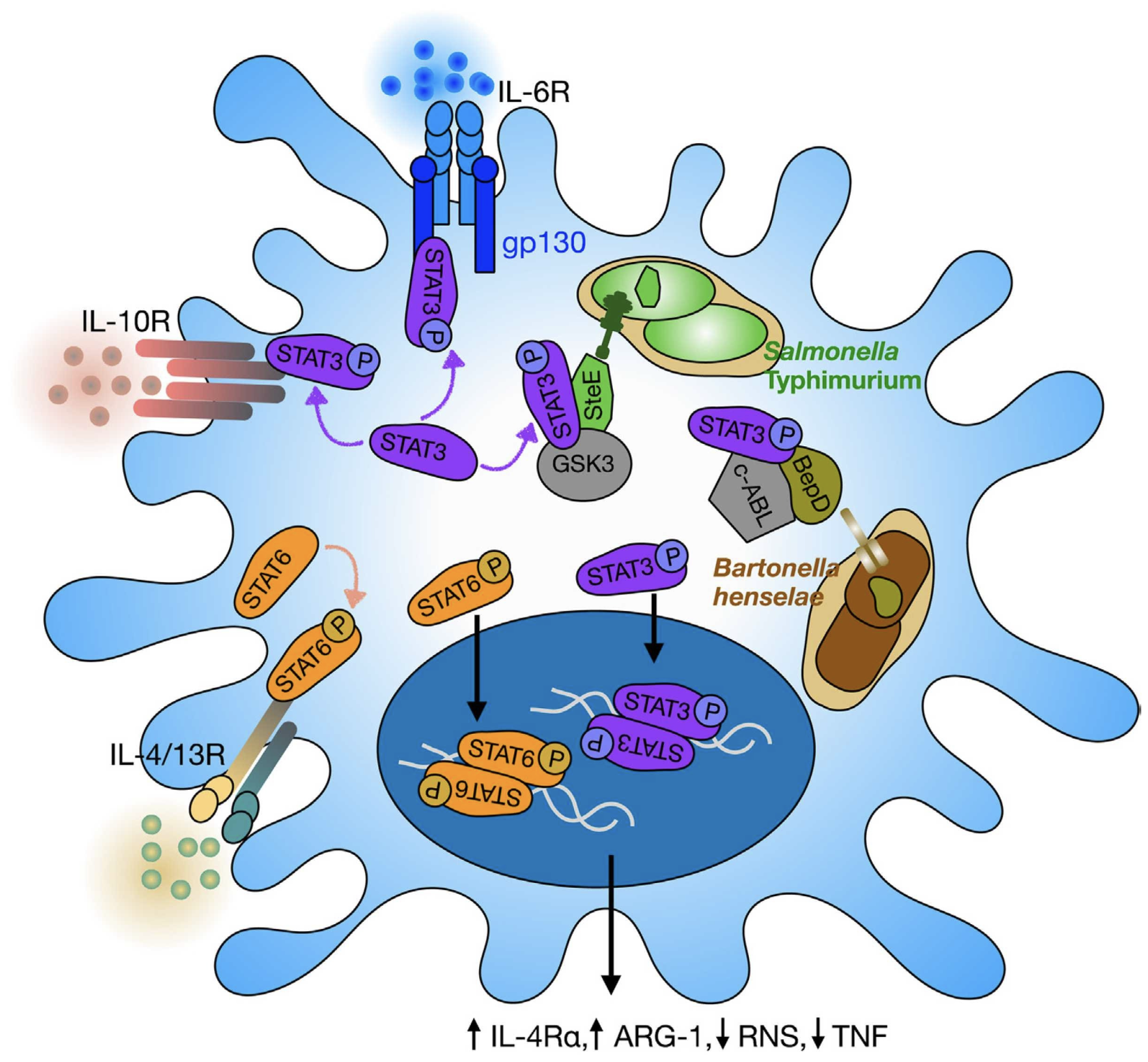
Turning foes into permissive hosts: manipulation of macrophage polarization by intracellular bacteria
Current Opinion in Immunology, 2023
Macrophages function as tissue-immune sentinels and mediate key antimicrobial responses against bacterial pathogens. Yet, they can also act as a cellular niche for intracellular bacteria, such as Salmonella enterica, to persist in infected tissues. Macrophages exhibit heterogeneous activation or polarization, states that are linked to differential antibacterial responses and bacteria permissiveness. Remarkably, recent studies demonstrate that Salmonella and other intracellular bacteria inject virulence effectors into the cellular cytoplasm to skew the macrophage polarization state and reprogram these immune cells into a permissive niche. Here, we review mechanisms of macrophage reprogramming by Salmonella and highlight manipulation of macrophage polarization as a shared bacterial pathogenesis strategy. In addition, we discuss how the interplay of bacterial effector mechanisms, microenvironmental signals, and ontogeny may shape macrophage cell states and functions. Finally, we propose ideas of how further research will advance our understanding of macrophage functional diversity and immunobiology.
Salmonella enterica serovar Typhi uses two type 3 secretion systems to replicate in human macrophages and colonize humanized mice
Salmonella enterica serovar Typhi (S. Typhi) is a human-restricted pathogen that replicates in macrophages. In this study, we investigated the roles of the S. Typhi type 3 secretion systems (T3SSs) encoded on Salmonella pathogenicity islands (SPI)-1 (T3SS-1) and SPI-2 (T3SS-2) during human macrophage infection. We found that mutants of S. Typhi deficient for both T3SSs were defective for intramacrophage replication as measured by flow cytometry, viable bacterial counts, and live time-lapse microscopy. T3SS-secreted proteins PipB2 and SifA contributed to S. Typhi replication and were translocated into the cytosol of human macrophages through both T3SS-1 and T3SS-2, demonstrating functional redundancy for these secretion systems. Importantly, an S. Typhi mutant strain that is deficient for both T3SS-1 and T3SS-2 was severely attenuated in the ability to colonize systemic tissues in a humanized mouse model of typhoid fever. Overall, this study establishes a critical role for S. Typhi T3SSs during its replication within human macrophages and during systemic infection of humanized mice.
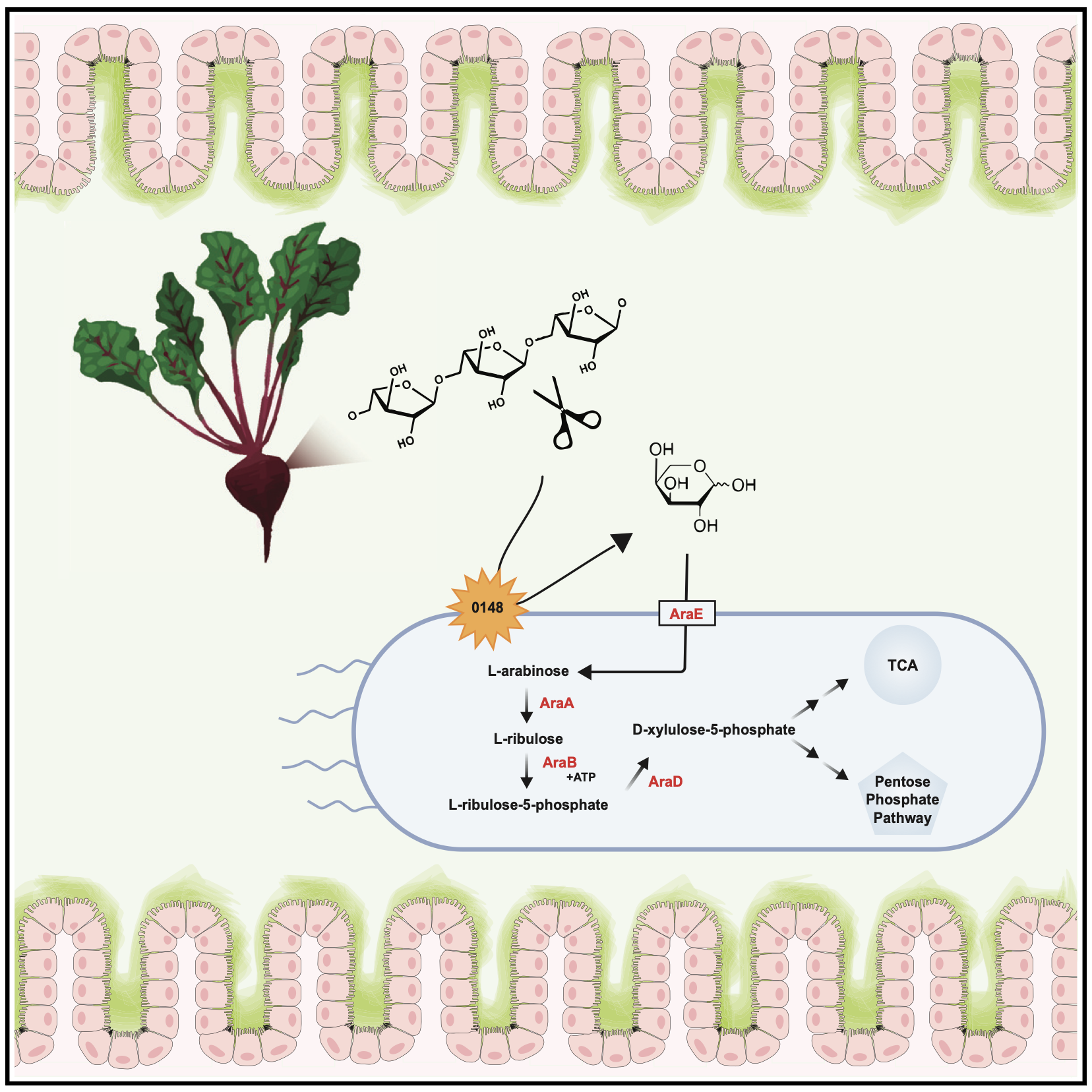
Salmonella-liberated dietary L-arabinose promotes expansion in superspreaders
The molecular understanding of host-pathogen interactions in the gastrointestinal (GI) tract of superspreader hosts is incomplete. In a mouse model of chronic, asymptomatic Salmonella enterica serovar Typhimurium (S. Tm) infection, we performed untargeted metabolomics on the feces of mice and found that superspreader hosts possess distinct metabolic signatures compared with non-superspreaders, including differential levels of L-arabinose. RNA-seq on S. Tm from superspreader fecal samples showed increased expression of the L-arabinose catabolism pathway in vivo. By combining bacterial genetics and diet manipulation, we demon- strate that diet-derived L-arabinose provides S. Tm a competitive advantage in the GI tract, and expansion of S. Tm in the GI tract requires an alpha-N-arabinofuranosidase that liberates L-arabinose from dietary poly- saccharides. Ultimately, our work shows that pathogen-liberated L-arabinose from the diet provides a competitive advantage to S. Tm in vivo. These findings propose L-arabinose as a critical driver of S. Tm expansion in the GI tracts of superspreader hosts.
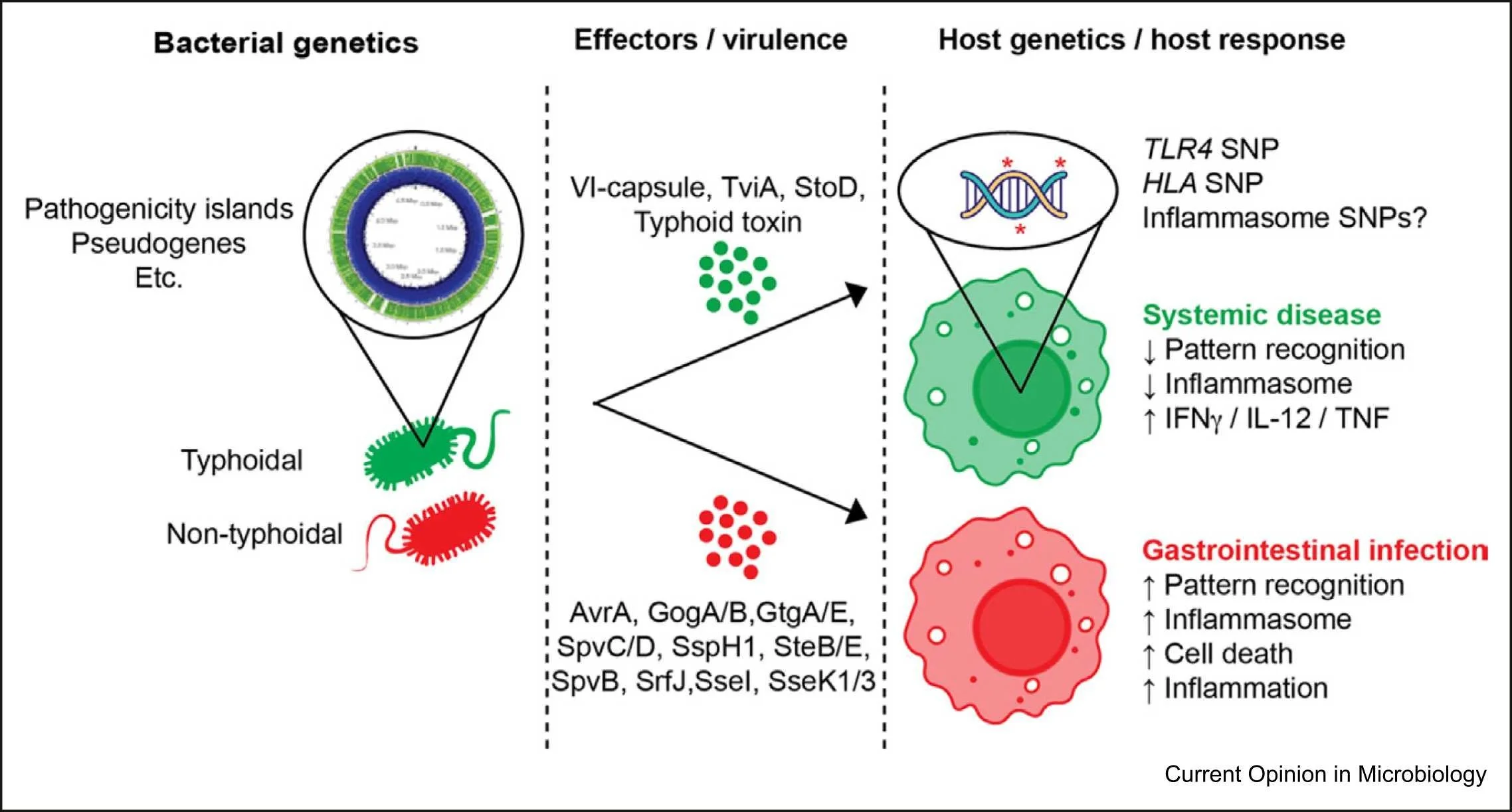
One species, different diseases: the unique molecular mechanisms that underlie the pathogenesis of typhoidal Salmonella infections
Current Opinion in Microbiology, 2023
Salmonella enterica is one of the most widespread bacterial pathogens found worldwide, resulting in approximately 100 million infections and over 200 000 deaths per year. Salmonella isolates, termed ‘serovars’, can largely be classified as either nontyphoidal or typhoidal Salmonella, which differ in regard to disease manifestation and host tropism. Nontyphoidal Salmonella causes gastroenteritis in many hosts, while typhoidal Salmonella is human-restricted and causes typhoid fever, a systemic disease with a mortality rate of up to 30% without treatment. There has been considerable interest in understanding how different Salmonella serovars cause different diseases, but the molecular details that underlie these infections have not yet been fully characterized, especially in the case of typhoidal Salmonella.In this review, we highlight the current state of research into understanding the pathogenesis of both nontyphoidal and typhoidal Salmonella, with a specific interest in serovar-specific traits that allow human-adapted strains of Salmonella to cause enteric fever. Overall, a more detailed molecular understanding of how different Salmonella isolates infect humans will provide critical insights into how we can eradicate these dangerous enteric pathogens.
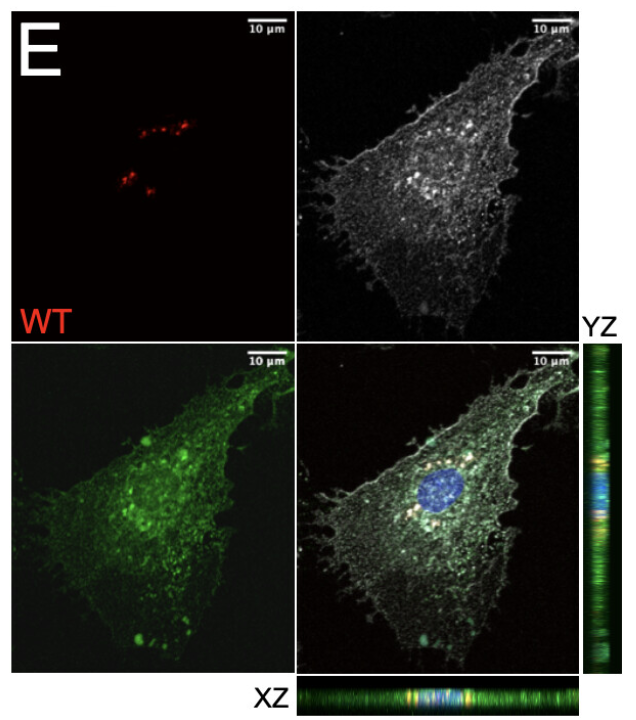
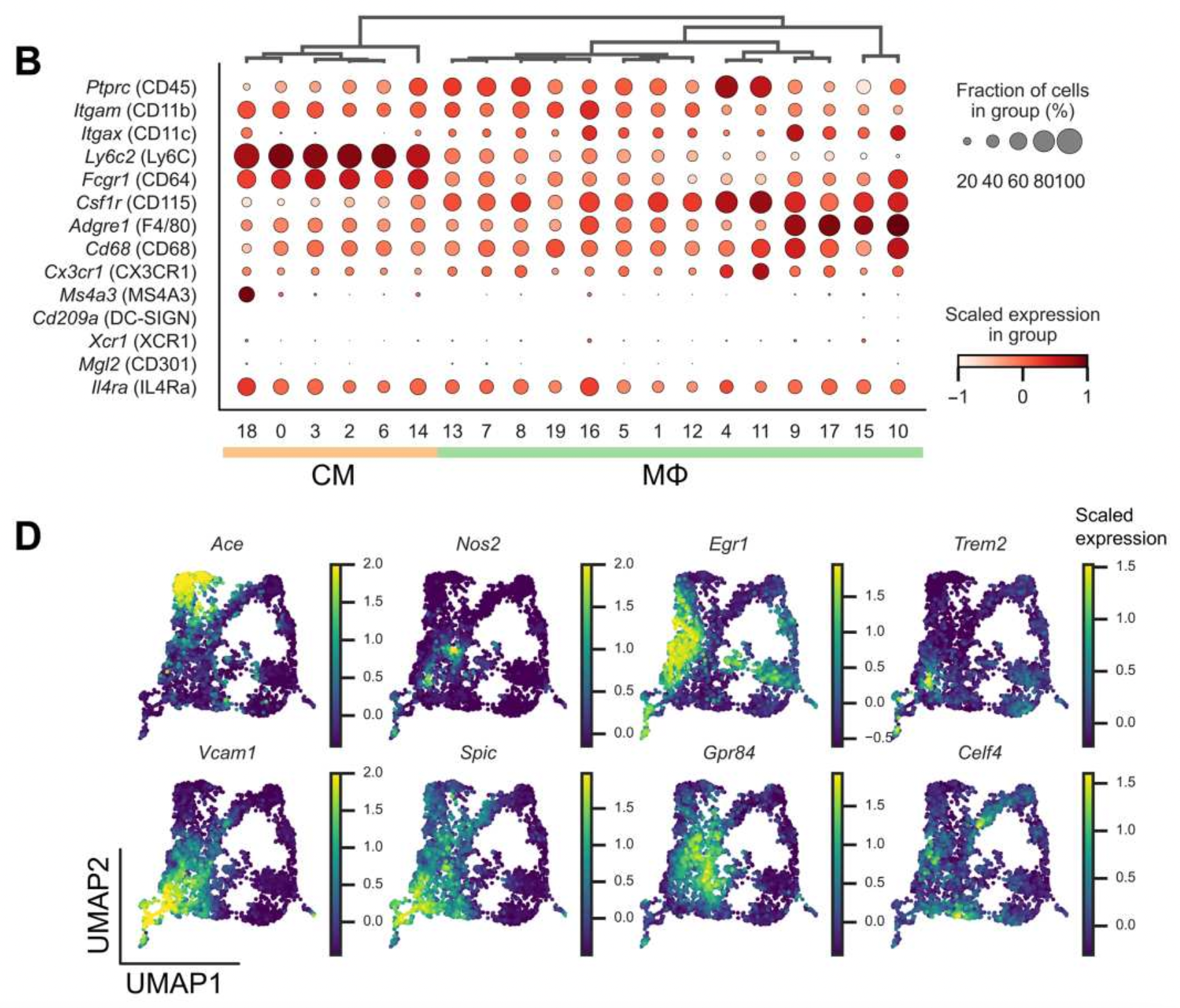
Single-cell profiling identifies ACE+ granuloma macrophages as a nonpermissive niche for intracellular bacteria during persistent Salmonella infection
Macrophages mediate key antimicrobial responses against intracellular bacterial pathogens, such as Salmonella enterica. Yet, they can also act as a permissive niche for these pathogens to persist in infected tissues within granulomas, which are immunological structures composed of macrophages and other immune cells. We apply single-cell transcriptomics to investigate macrophage functional diversity during persistent S. enterica serovar Typhimurium (STm) infection in mice. We identify determinants of macrophage heterogeneity in infected spleens and describe populations of distinct phenotypes, functional programming, and spatial localization. Using an STm mutant with impaired ability to polarize macrophage phenotypes, we find that angiotensin-converting enzyme (ACE) defines a granuloma macrophage population that is nonpermissive for intracellular bacteria, and their abundance anticorrelates with tissue bacterial burden. Disruption of pathogen control by neutralizing TNF is linked to preferential depletion of ACE+macrophages in infected tissues. Thus, ACE+ macrophages have limited capacity to serve as cellular niche for intracellular bacteria to establish persistent infection.
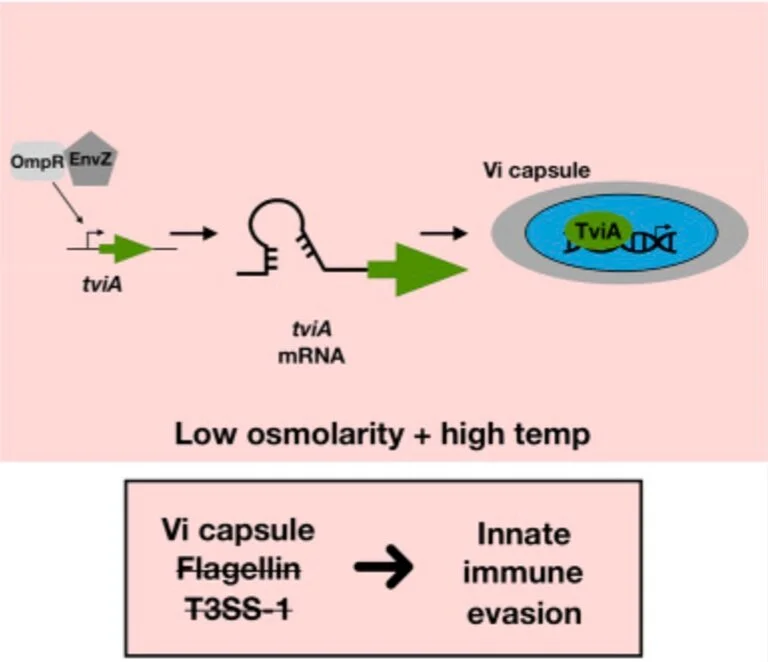
A Salmonella Typhi RNA thermosensor regulates virulence factors and innate immune evasion in response to host temperature
Sensing and responding to environmental signals is critical for bacterial pathogens to successfully infect and persist within hosts. Many bacterial pathogens sense temperature as an indication they have entered a new host and must alter their virulence factor expression to evade immune detection. Using secondary structure prediction, we identified an RNA thermosensor (RNAT) in the 5' untranslated region (UTR) of tviA encoded by the typhoid fever-causing bacterium Salmonella enterica serovar Typhi (S. Typhi). Importantly, tviA is a transcriptional regulator of the critical virulence factors Vi capsule, flagellin, and type III secretion system-1 expression. By introducing point mutations to alter the mRNA secondary structure, we demonstrate that the 5' UTR of tviA contains a functional RNAT using in vitro expression, structure probing, and ribosome binding methods. Mutational inhibition of the RNAT in S. Typhi causes aberrant virulence factor expression, leading to enhanced innate immune responses during infection. In conclusion, we show that S. Typhi regulates virulence factor expression through an RNAT in the 5' UTR of tviA. Our findings demonstrate that limiting inflammation through RNAT-dependent regulation in response to host body temperature is important for S. Typhi's "stealthy" pathogenesis.
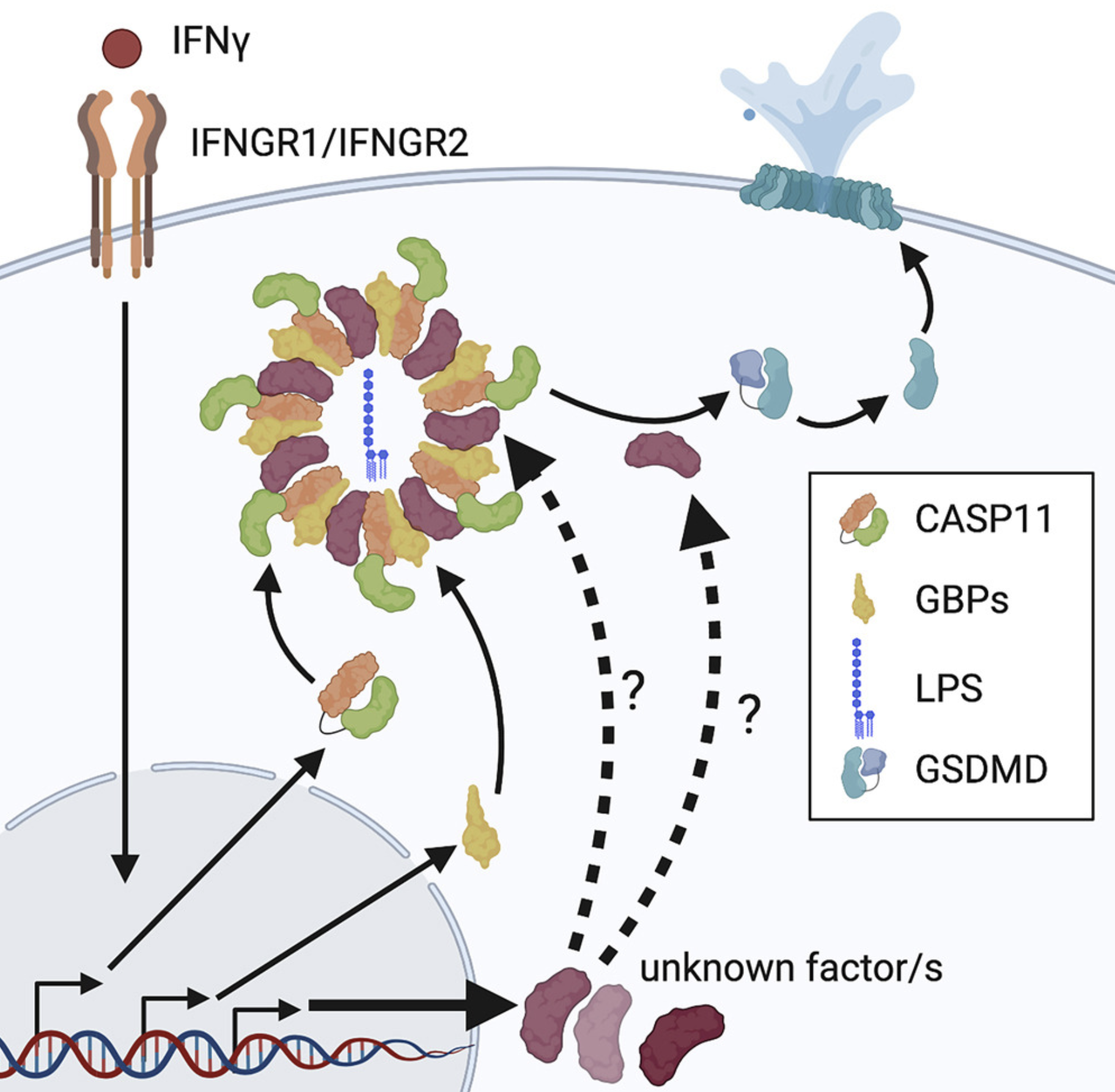
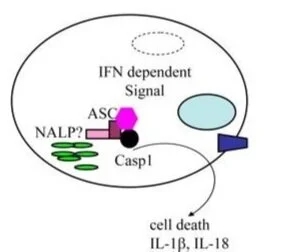
A Rapid Caspase-11 Response Induced by IFN γ Priming Is Independent of Guanylate Binding Proteins
In mammalian cells, inflammatory caspases detect Gram-negative bacterial invasion by binding lipopolysaccharides (LPS). Murine caspase-11 binds cytosolic LPS, stimulates pyroptotic cell death, and drives sepsis pathogenesis. Extracellular priming factors enhance caspase-11-dependent pyroptosis. Herein we compare priming agents and demonstrate that IFNγ priming elicits the most rapid and amplified macrophage response to cytosolic LPS. Previous studies indicate that IFN-induced expression of caspase-11 and guanylate binding proteins (GBPs) are causal events explaining the effects of priming on cytosolic LPS sensing. We demonstrate that these events cannot fully account for the increased response triggered by IFNγ treatment. Indeed, IFNγ priming elicits higher pyroptosis levels in response to cytosolic LPS when macrophages stably express caspase-11. In macrophages lacking GBPs encoded on chromosome 3, IFNγ priming enhanced pyroptosis in response to cytosolic LPS as compared with other priming agents. These results suggest an unknown regulator of caspase-11-dependent pyroptosis exists, whose activity is upregulated by IFNγ.
Genetic variation in the MacAB-TolC efflux pump influences pathogenesis of invasive Salmonella isolates from Africa
The various sub-species of Salmonella enterica cause a range of disease in human hosts. The human-adapted Salmonella enterica serovar Typhi enters the gastrointestinal tract and invades systemic sites to cause enteric (typhoid) fever. In contrast, most non-typhoidal serovars of Salmonella are primarily restricted to gut tissues. Across Africa, invasive non-typhoidal Salmonella (iNTS) have emerged with an ability to spread beyond the gastrointestinal tract and cause systemic bloodstream infections with increased morbidity and mortality. To investigate this evolution in pathogenesis, we compared the genomes of African iNTS isolates with other Salmonella enterica serovar Typhimurium and identified several macA and macB gene variants unique to African iNTS. MacAB forms a tripartite efflux pump with TolC and is implicated in Salmonella pathogenesis. We show that macAB transcription is upregulated during macrophage infection and after antimicrobial peptide exposure, with macAB transcription being supported by the PhoP/Q two-component system. Constitutive expression of macAB improves survival of Salmonella in the presence of the antimicrobial peptide C18G. Furthermore, these macAB variants affect replication in macrophages and influence fitness during colonization of the murine gastrointestinal tract. Importantly, the infection outcome resulting from these macAB variants depends upon both the Salmonella Typhimurium genetic background and the host gene Nramp1, an important determinant of innate resistance to intracellular bacterial infection. The variations we have identified in the MacAB-TolC efflux pump in African iNTS may reflect evolution within human host populations that are compromised in their ability to clear intracellular Salmonella infections.
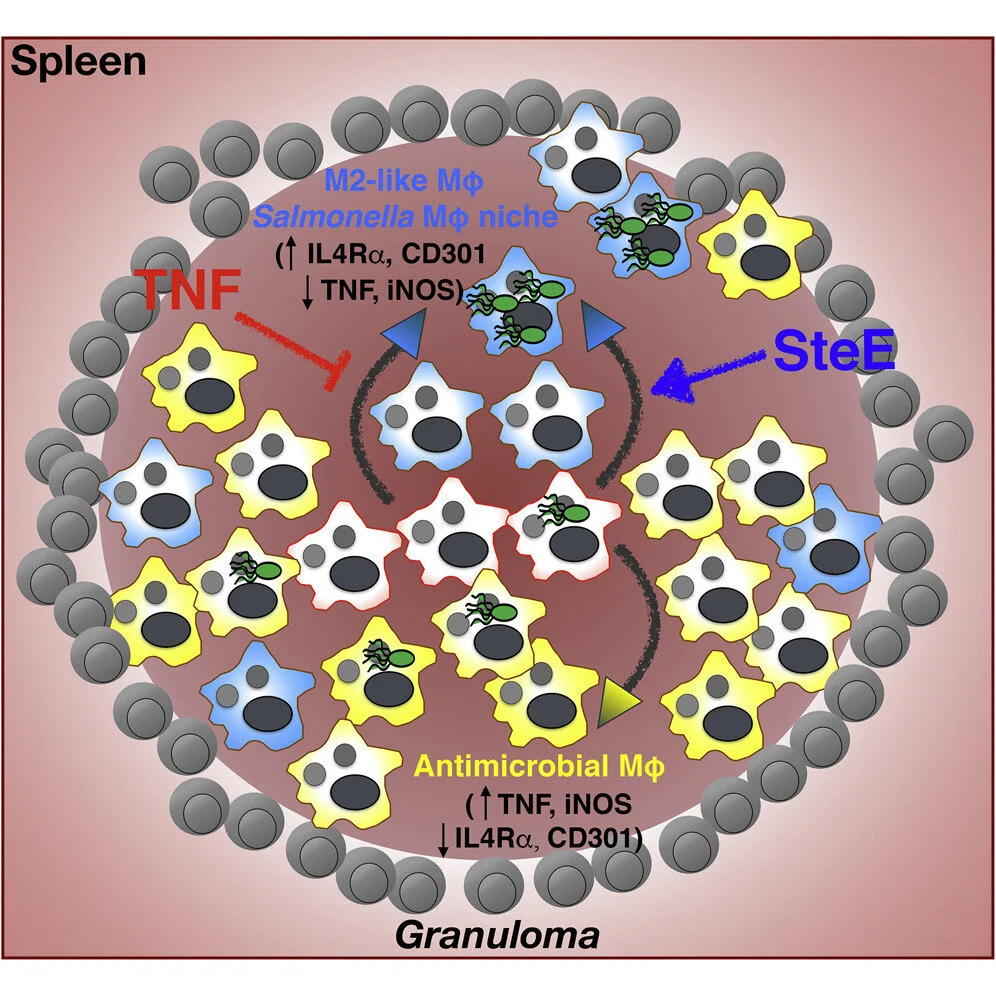
Salmonella-Driven Polarization of Granuloma Macrophages Antagonizes TNF-Mediated Pathogen Restriction during Persistent Infection
Many intracellular bacteria can establish chronic infection and persist in tissues within granulomas composed of macrophages. Granuloma macrophages exhibit heterogeneous polarization states, or phenotypes, that may be functionally distinct. Here, we elucidate a host-pathogen interaction that controls granuloma macrophage polarization and long-term pathogen persistence during Salmonella Typhimurium (STm) infection. We show that STm persists within splenic granulomas that are densely populated by CD11b+CD11c+Ly6C+ macrophages. STm preferentially persists in granuloma macrophages reprogrammed to an M2 state, in part through the activity of the effector SteE, which contributes to the establishment of persistent infection. We demonstrate that tumor necrosis factor (TNF) signaling limits M2 granuloma macrophage polarization, thereby restricting STm persistence. TNF neutralization shifts granuloma macrophages toward an M2 state and increases bacterial persistence, and these effects are partially dependent on SteE activity. Thus, manipulating granuloma macrophage polarization represents a strategy for intracellular bacteria to overcome host restriction during persistent infection.
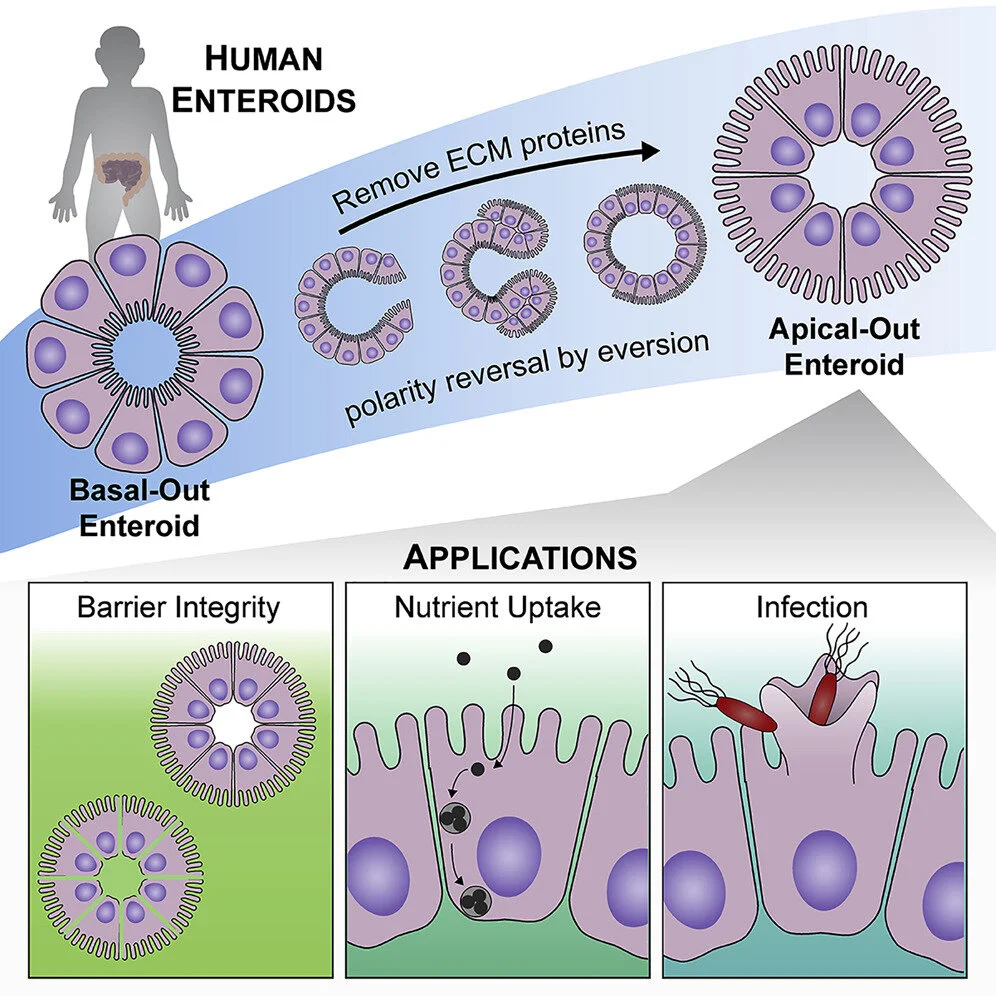
Controlling Epithelial Polarity: A Human Enteroid Model for Host-Pathogen Interactions
Human enteroids—epithelial spheroids derived from primary gastrointestinal tissue—are a promising model to study pathogen-epithelial interactions. However, accessing the apical enteroid surface is challenging because it is enclosed within the spheroid. We developed a technique to reverse enteroid polarity such that the apical surface everts to face the media. Apical-out enteroids maintain proper polarity and barrier function, differentiate into the major intestinal epithelial cell (IEC) types, and exhibit polarized absorption of nutrients. We used this model to study host-pathogen interactions and identified distinct polarity-specific patterns of infection by invasive enteropathogens. Salmonella enterica serovar Typhimurium targets IEC apical surfaces for invasion via cytoskeletal rearrangements, and Listeria monocytogenes, which binds to basolateral receptors, invade apical surfaces at sites of cell extrusion. Despite different modes of entry, both pathogens exit the epithelium within apically extruding enteroid cells. This model will enable further examination of IECs in health and disease.
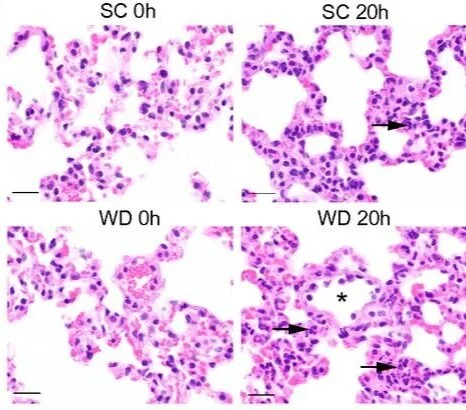
Western diet regulates immune status and the response to LPS-driven sepsis independent of diet-associated microbiome
Proceedings of the National Academy of Sciences, 2019
The Western diet (WD) is high in fats and sucrose and low in fiber and is the most prevalent diet in westernized countries. We find that in our model of sepsis, mice fed WD had increased sepsis severity and poorer outcomes. WD-fed mice had higher baseline inflammation, increased sepsis-associated immunoparalysis, and altered neutrophil populations in the blood. The WD-dependent increase in sepsis severity and mortality was independent of the diet-associated microbiome, suggesting that diet may be directly regulating innate immunity. We used our identified disease factors and found WD-fed mice occupy a unique path in sepsis disease progression. Our data provide insight into diet-dependent reprogramming of the immune response and will be important in treating and diagnosing a WD-fed population.
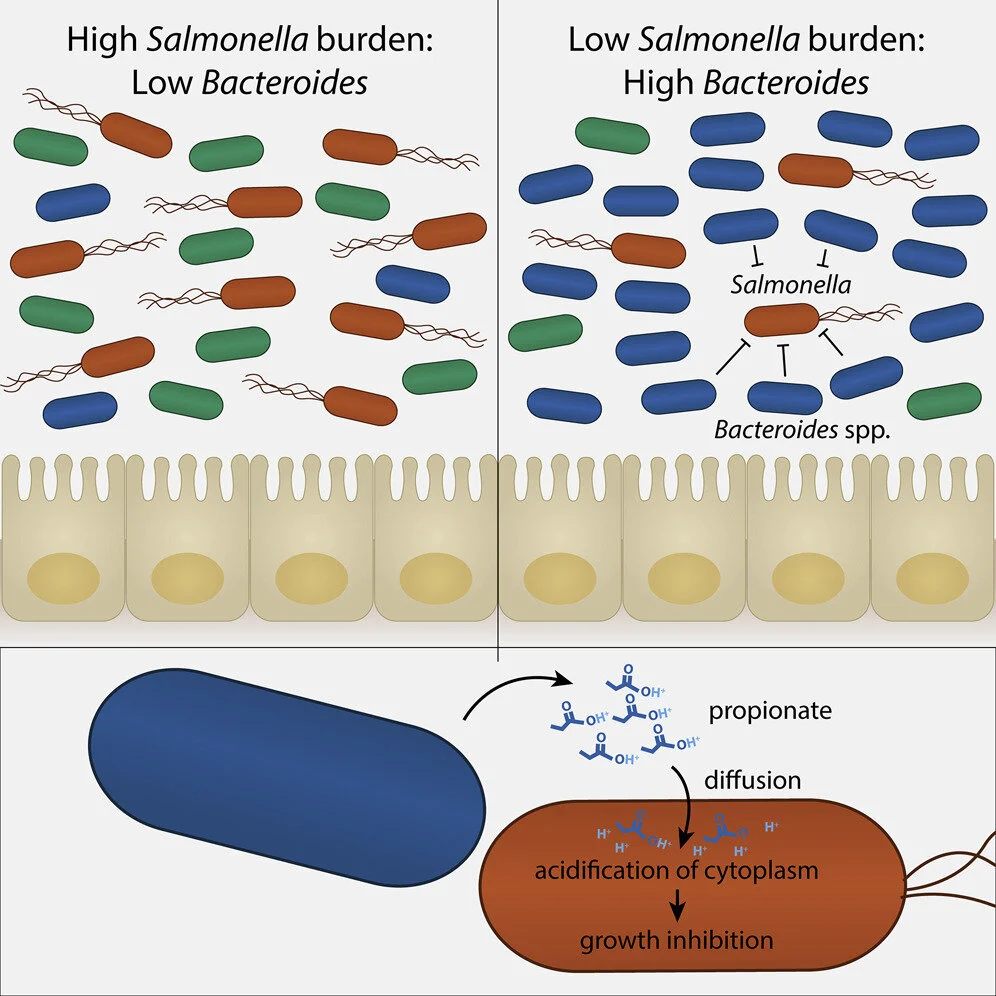
A Gut Commensal-Produced Metabolite Mediates Colonization Resistance to Salmonella Infection
The intestinal microbiota provides colonization resistance against pathogens, limiting pathogen expansion and transmission. These microbiota-mediated mechanisms were previously identified by observing loss of colonization resistance after antibiotic treatment or dietary changes, which severely disrupt microbiota communities. We identify a microbiota-mediated mechanism of colonization resistance against Salmonella enterica serovar Typhimurium (S. Typhimurium) by comparing high-complexity commensal communities with different levels of colonization resistance. Using inbred mouse strains with different infection dynamics and S. Typhimurium intestinal burdens, we demonstrate that Bacteroides species mediate colonization resistance against S. Typhimurium by producing the short-chain fatty acid propionate. Propionate directly inhibits pathogen growth in vitro by disrupting intracellular pH homeostasis, and chemically increasing intestinal propionate levels protects mice from S. Typhimurium. In addition, administering susceptible mice Bacteroides, but not a propionate-production mutant, confers resistance to S. Typhimurium. This work provides mechanistic understanding into the role of individualized microbial communities in host-to-host variability of pathogen transmission.
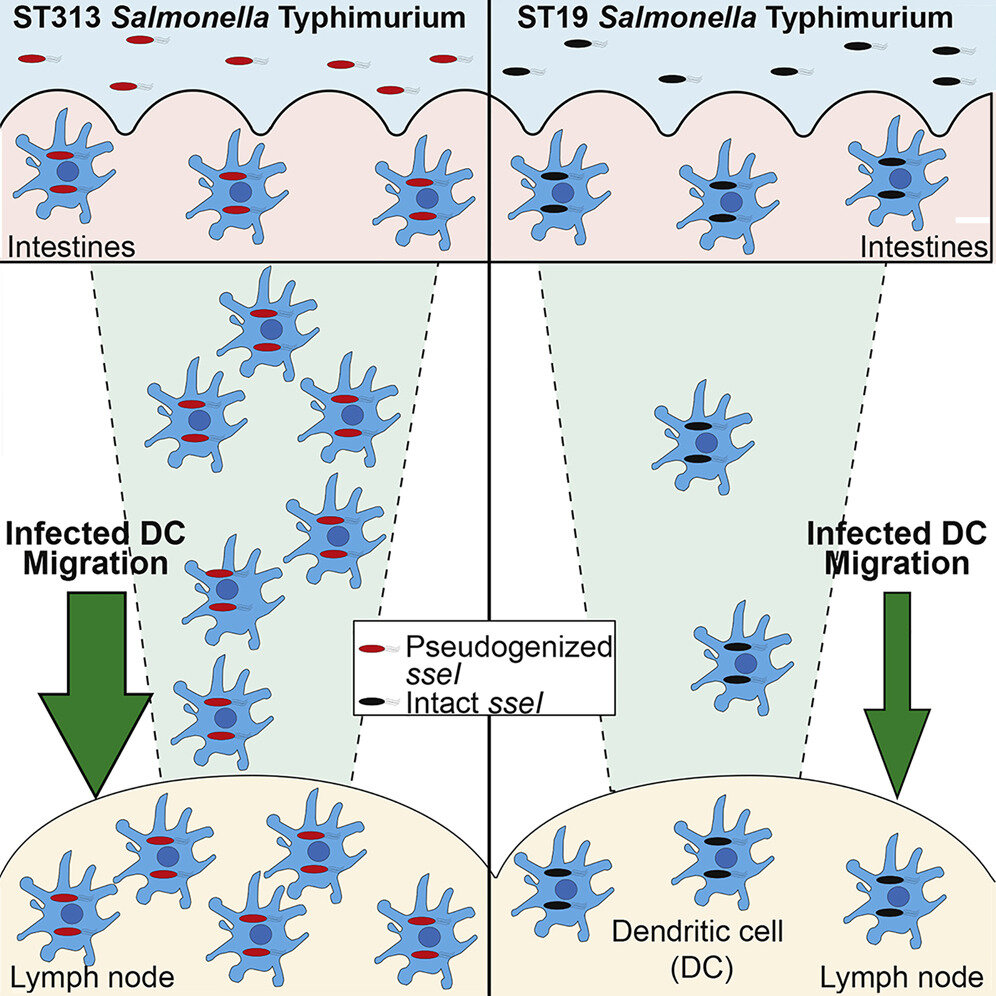
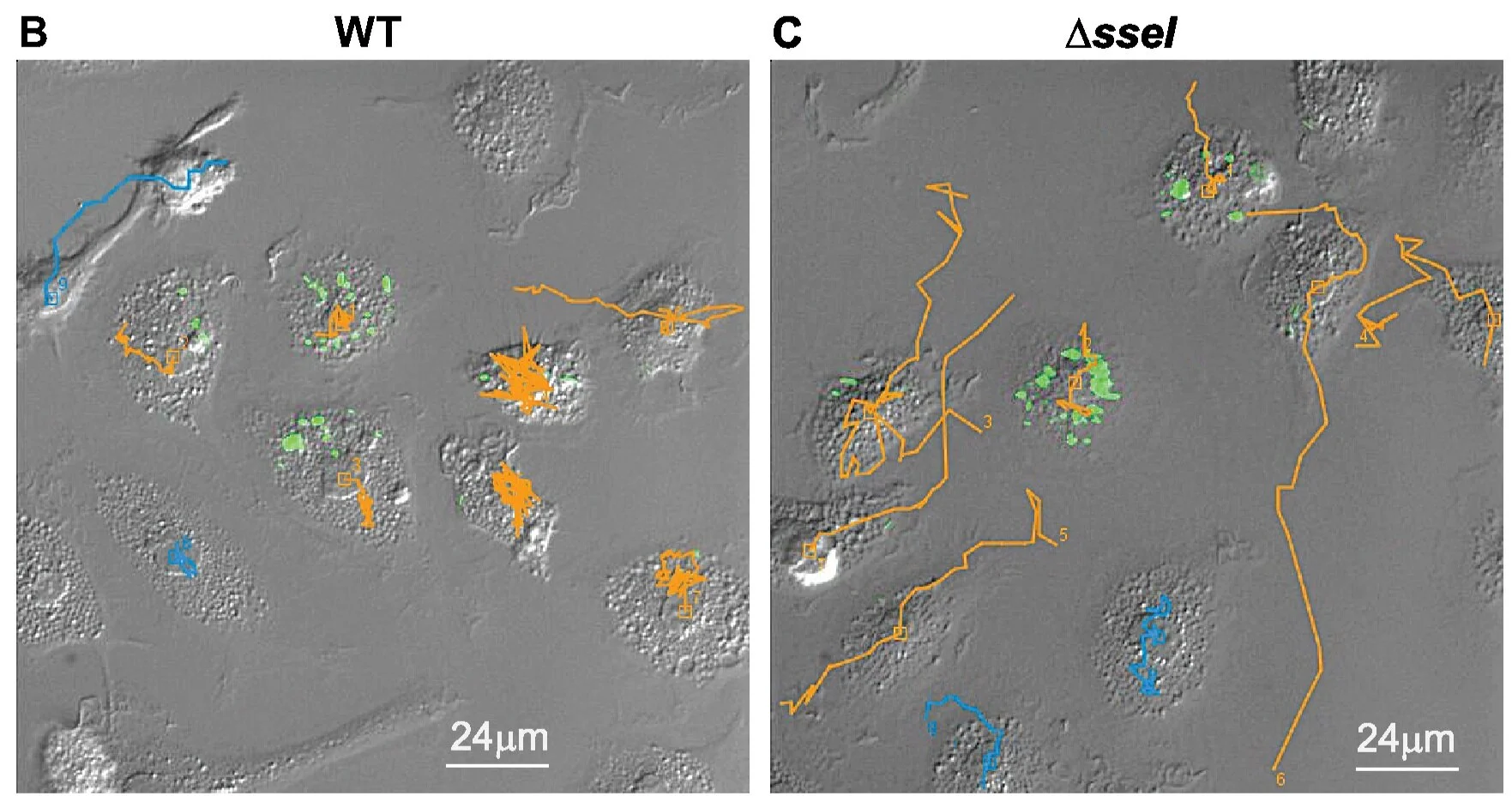
Pseudogenization of the Secreted Effector Gene sseI Confers Rapid Systemic Dissemination of S. Typhimurium ST313 within Migratory Dendritic Cells
Genome degradation correlates with host adaptation and systemic disease in Salmonella. Most lineages of the S. enterica subspecies Typhimurium cause gastroenteritis in humans; however, the recently emerged ST313 lineage II pathovar commonly causes systemic bacteremia in sub-Saharan Africa. ST313 lineage II displays genome degradation compared to gastroenteritis-associated lineages; yet, the mechanisms and causal genetic differences mediating these infection phenotypes are largely unknown. We find that the ST313 isolate D23580 hyperdisseminates from the gut to systemic sites, such as the mesenteric lymph nodes (MLNs), via CD11b+ migratory dendritic cells (DCs). This hyperdissemination was facilitated by the loss of sseI, which encodes an effector that inhibits DC migration in gastroenteritis-associated isolates. Expressing functional SseI in D23580 reduced the number of infected migratory DCs and bacteria in the MLN. Our study reveals a mechanism linking pseudogenization of effectors with the evolution of niche adaptation in a bacterial pathogen.
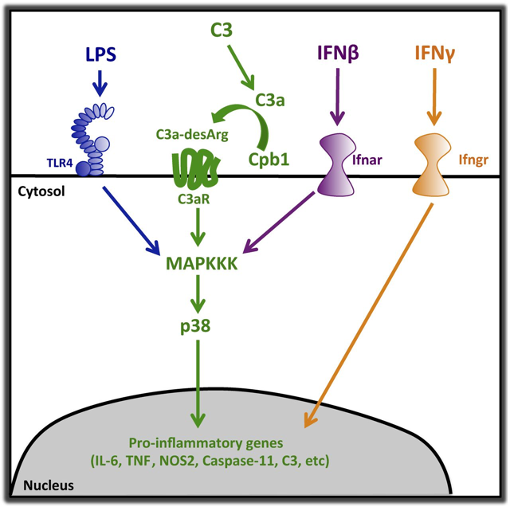
Complement pathway amplifies caspase-11–dependent cell death and endotoxin-induced sepsis severity
Journal of Experimental Medicine, 2016
Cell death and release of proinflammatory mediators contribute to mortality during sepsis. Specifically, caspase-11–dependent cell death contributes to pathology and decreases in survival time in sepsis models. Priming of the host cell, through TLR4 and interferon receptors, induces caspase-11 expression, and cytosolic LPS directly stimulates caspase-11 activation, promoting the release of proinflammatory cytokines through pyroptosis and caspase-1 activation. Using a CRISPR-Cas9–mediated genome-wide screen, we identified novel mediators of caspase-11–dependent cell death. We found a complement-related peptidase, carboxypeptidase B1 (Cpb1), to be required for caspase-11 gene expression and subsequent caspase-11–dependent cell death. Cpb1 modifies a cleavage product of C3, which binds to and activates C3aR, and then modulates innate immune signaling. We find the Cpb1–C3–C3aR pathway induces caspase-11 expression through amplification of MAPK activity downstream of TLR4 and Ifnar activation, and mediates severity of LPS-induced sepsis (endotoxemia) and disease outcome in mice. We show C3aR is required for up-regulation of caspase-11 orthologues, caspase-4 and -5, in primary human macrophages during inflammation and that c3aR1 and caspase-5 transcripts are highly expressed in patients with severe sepsis; thus, suggesting that these pathways are important in human sepsis. Our results highlight a novel role for complement and the Cpb1–C3–C3aR pathway in proinflammatory signaling, caspase-11 cell death, and sepsis severity.
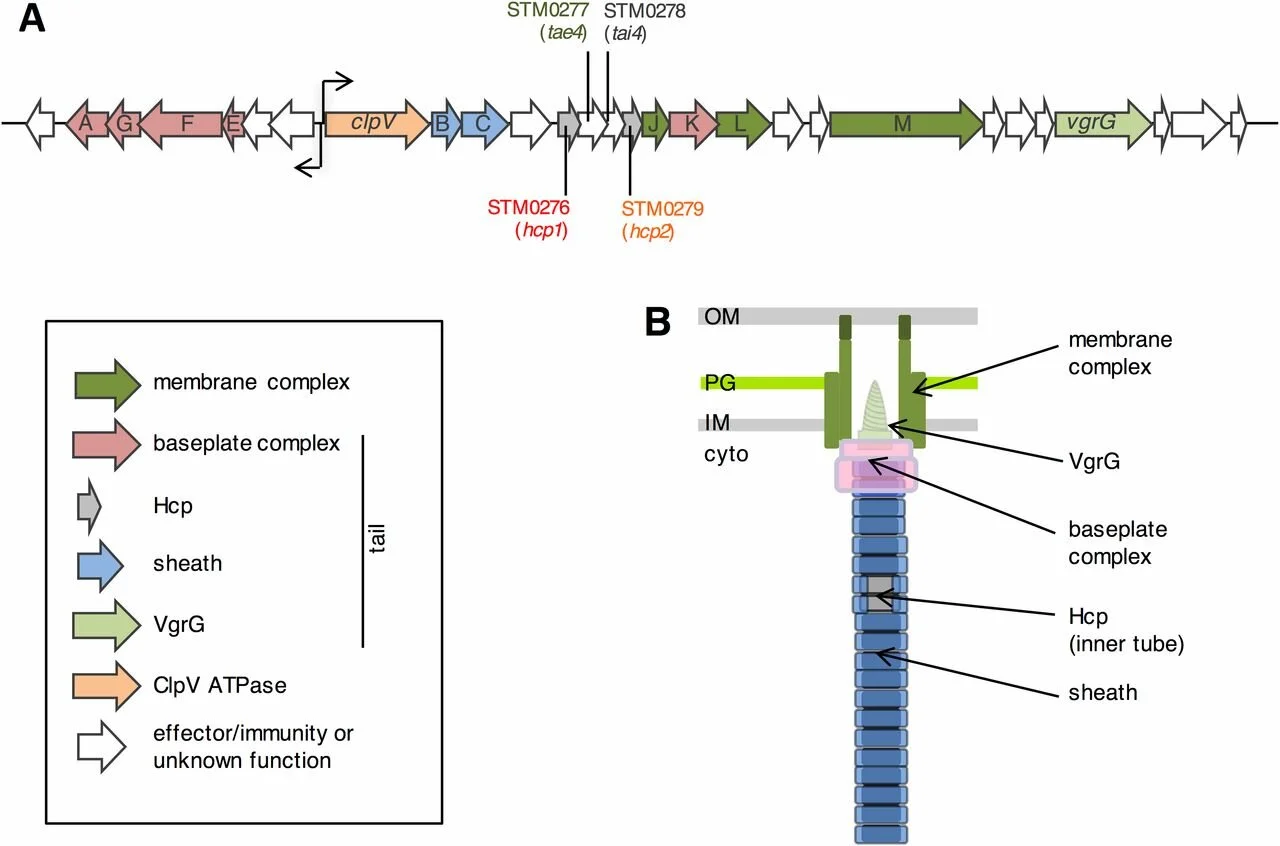
Salmonella Typhimurium utilizes a T6SS-mediated antibacterial weapon to establish in the host gut
Proceedings of the National Academy of Sciences, 2016
Gram-negative bacteria use the type VI secretion system (T6SS) to deliver effectors into adjacent cells. Salmonella Typhimurium is an enteric pathogen that causes disease in millions of individuals each year. Its ability to infect the mammalian gut is a key factor that contributes to its virulence and transmission to new hosts. However, many of the details on how Salmonella successfully colonizes the gut and persists among members of the gut microbiota remain to be deciphered. In this work, we provide evidence that Salmonella uses an antibacterial weapon, the type VI secretion system, to establish infection in the gut. In addition, our results suggest that S. Typhimurium selectively targets specific members of the microbiota to invade the gastrointestinal tract.
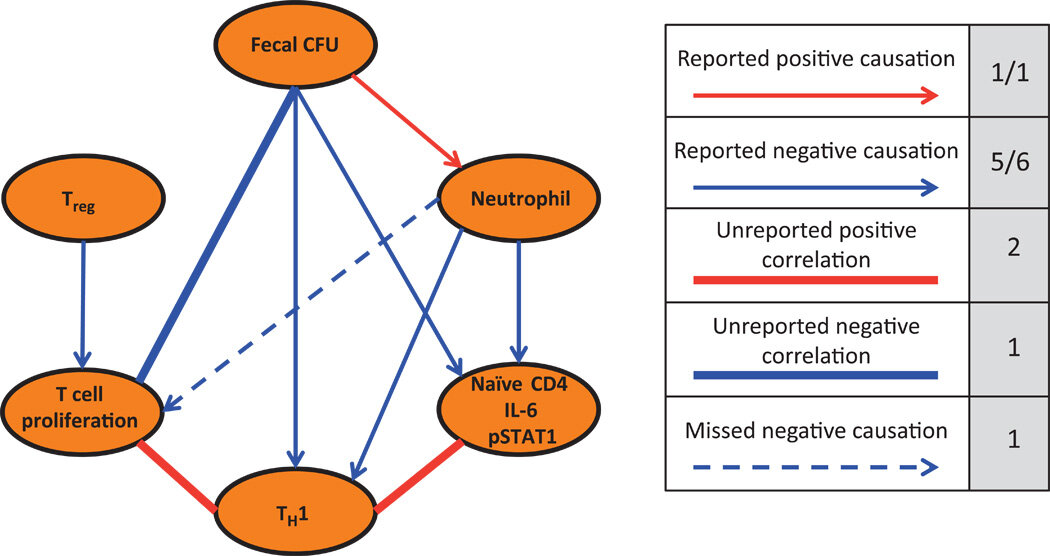
Coordinate actions of innate immune responses oppose those of the adaptive immune system during Salmonella infection of mice
The immune system enacts a coordinated response when faced with complex environmental and pathogenic perturbations. We used the heterogeneous responses of mice to persistent Salmonella infection to model system-wide coordination of the immune response to bacterial burden. We hypothesized that the variability in outcomes of bacterial growth and immune response across genetically identical mice could be used to identify immune elements that serve as integrators enabling co-regulation and interconnectedness of the innate and adaptive immune systems. Correlation analysis of immune response variation to Salmonella infection linked bacterial load with at least four discrete, interacting functional immune response “cassettes.” One of these, the innate cassette, in the chronically infected mice included features of the innate immune system, systemic neutrophilia, and high serum concentrations of the proinflammatory cytokine interleukin-6. Compared with mice with a moderate bacterial load, mice with the highest bacterial burden exhibited high activity of this innate cassette, which was associated with a dampened activity of the adaptive T cell cassette—with fewer plasma cells and CD4+ T helper 1 cells and increased numbers of regulatory T cells—and with a dampened activity of the cytokine signaling cassette. System-wide manipulation of neutrophil numbers revealed that neutrophils regulated signal transducer and activator of transcription (STAT) signaling in B cells during infection. Thus, a network-level approach demonstrated unappreciated interconnections that balanced innate and adaptive immune responses during the dynamic course of disease and identified signals associated with pathogen transmission status, as well as a regulatory role for neutrophils in cytokine signaling.
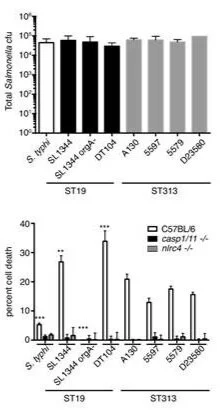
Non-typhoidal Salmonella Typhimurium ST313 isolates that cause bacteremia in humans stimulate less inflammasome activation than ST19 isolates associated with gastroenteritis
Salmonella is an enteric pathogen that causes a range of diseases in humans. Non-typhoidal Salmonella (NTS) serovars such as Salmonella enterica serovar Typhimurium generally cause a self-limiting gastroenteritis whereas typhoidal serovars cause a systemic disease, typhoid fever. However, S. Typhimurium isolates within the multi-locus sequence type ST313 have emerged in sub-Saharan Africa as a major cause of bacteremia in humans. The S. Typhimurium ST313 lineage is phylogenetically distinct from classical S. Typhimurium lineages, such as ST19, that cause zoonotic gastroenteritis worldwide. Previous studies have shown that the ST313 lineage has undergone genome degradation when compared to the ST19 lineage, similar to that observed for typhoidal serovars. Currently, little is known about phenotypic differences between ST313 isolates and other NTS isolates. We find that representative ST313 isolates invade non-phagocytic cells less efficiently than the classical ST19 isolates that are more commonly associated with gastroenteritis. In addition, ST313 isolates induce less Caspase-1-dependent macrophage death and IL-1 release than ST19 isolates. ST313 isolates also express relatively lower levels of mRNA of the genes encoding the SPI-1 effector sopE2 and the flagellin, fliC, providing possible explanations for the decrease in invasion and inflammasome activation. The ST313 isolates have invasion and inflammatory phenotypes that are intermediate; more invasive and inflammatory than Salmonella enterica serovar Typhi and less than ST19 isolates associated with gastroenteritis. This suggests that both phenotypically and at the genomic level ST313 isolates are evolving signatures that facilitate a systemic lifestyle in humans.
cGAS and Ifi204 cooperate to produce type I IFNs in response to Francisella infection
Type I IFN production is an important host immune response against viral and bacterial infections. However, little is known about the ligands and corresponding host receptors that trigger type I IFN production during bacterial infections. We used a model intracellular pathogen, Francisella novicida, to begin characterizing the type I IFN response to bacterial pathogens. F. novicida replicates in the cytosol of host cells and elicits a robust type I IFN response that is largely TLR independent, but is dependent on the adapter molecule STING, suggesting that the type I IFN stimulus during F. novicida infection is cytosolic. In this study, we report that the cytosolic DNA sensors, cyclic GMP-AMP synthase (cGAS) and Ifi204, are both required for the STING-dependent type I IFN response to F. novicida infection in both primary and immortalized murine macrophages. We created cGAS, Ifi204, and Sting functional knockouts in RAW264.7 macrophages and demonstrated that cGAS and Ifi204 cooperate to sense dsDNA and activate the STING-dependent type I IFN pathway. In addition, we show that dsDNA from F. novicida is an important type I IFN stimulating ligand. One outcome of cGAS–STING signaling is the activation of the absent in melanoma 2 inflammasome in response to F. novicida infection. Whereas the absent in melanoma 2 inflammasome is beneficial to the host during F. novicida infection, type I IFN signaling by STING and IFN regulatory factor 3 is detrimental to the host during F. novicida infection. Collectively, our studies indicate that cGAS and Ifi204 cooperate to sense cytosolic dsDNA and F. novicida infection to produce a strong type I IFN response.
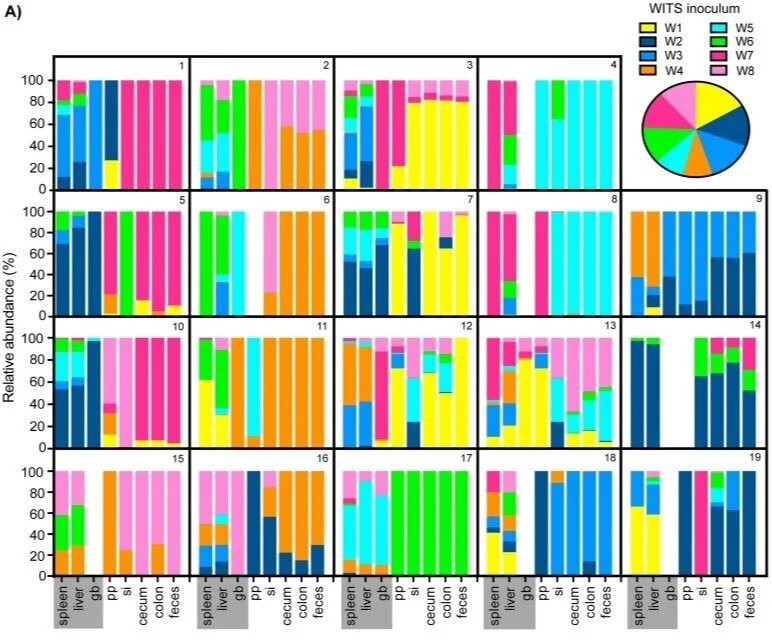
Intraspecies competition for niches in the distal gut dictate transmission during persistent Salmonella infection
In order to be transmitted, a pathogen must first successfully colonize and multiply within a host. Ecological principles can be applied to study host-pathogen interactions to predict transmission dynamics. Little is known about the population biology of Salmonella during persistent infection. To define Salmonella enterica serovar Typhimurium population structure in this context, 129SvJ mice were oral gavaged with a mixture of eight wild-type isogenic tagged Salmonella (WITS) strains. Distinct subpopulations arose within intestinal and systemic tissues after 35 days, and clonal expansion of the cecal and colonic subpopulation was responsible for increases in Salmonella fecal shedding. A co-infection system utilizing differentially marked isogenic strains was developed in which each mouse received one strain orally and the other systemically by intraperitoneal (IP) injection. Co-infections demonstrated that the intestinal subpopulation exerted intraspecies priority effects by excluding systemic S. Typhimurium from colonizing an extracellular niche within the cecum and colon. Importantly, the systemic strain was excluded from these distal gut sites and was not transmitted to naïve hosts. In addition, S. Typhimurium required hydrogenase, an enzyme that mediates acquisition of hydrogen from the gut microbiota, during the first week of infection to exert priority effects in the gut. Thus, early inhibitory priority effects are facilitated by the acquisition of nutrients, which allow S. Typhimurium to successfully compete for a nutritional niche in the distal gut. We also show that intraspecies colonization resistance is maintained by Salmonella Pathogenicity Islands SPI1 and SPI2 during persistent distal gut infection. Thus, important virulence effectors not only modulate interactions with host cells, but are crucial for Salmonella colonization of an extracellular intestinal niche and thereby also shape intraspecies dynamics. We conclude that priority effects and intraspecies competition for colonization niches in the distal gut control Salmonella population assembly and transmission.
Role of disease-associated tolerance in infectious superspreaders
Proceedings of the National Academy of Sciences, 2014
Natural populations show striking heterogeneity in their ability to transmit disease. For example, a minority of infected individuals known as superspreaders carries out the majority of pathogen transmission events. In a mouse model of Salmonella infection, a subset of infected hosts becomes superspreaders, shedding high levels of bacteria (>10^8 cfu per g of feces) but remain asymptomatic with a dampened systemic immune state. Here we show that superspreader hosts remain asymptomatic when they are treated with oral antibiotics. In contrast, nonsuperspreader Salmonella-infected hosts that are treated with oral antibiotics rapidly shed superspreader levels of the pathogen but display signs of morbidity. This morbidity is linked to an increase in inflammatory myeloid cells in the spleen followed by increased production of acute-phase proteins and proinflammatory cytokines. The degree of colonic inflammation is similar in antibiotic-treated superspreader and nonsuperspreader hosts, indicating that the superspreader hosts are tolerant of antibiotic-mediated perturbations in the intestinal tract. Importantly, neutralization of acute-phase proinflammatory cytokines in antibiotic-induced superspreaders suppresses the expansion of inflammatory myeloid cells and reduces morbidity. We describe a unique disease-associated tolerance to oral antibiotics in superspreaders that facilitates continued transmission of the pathogen.
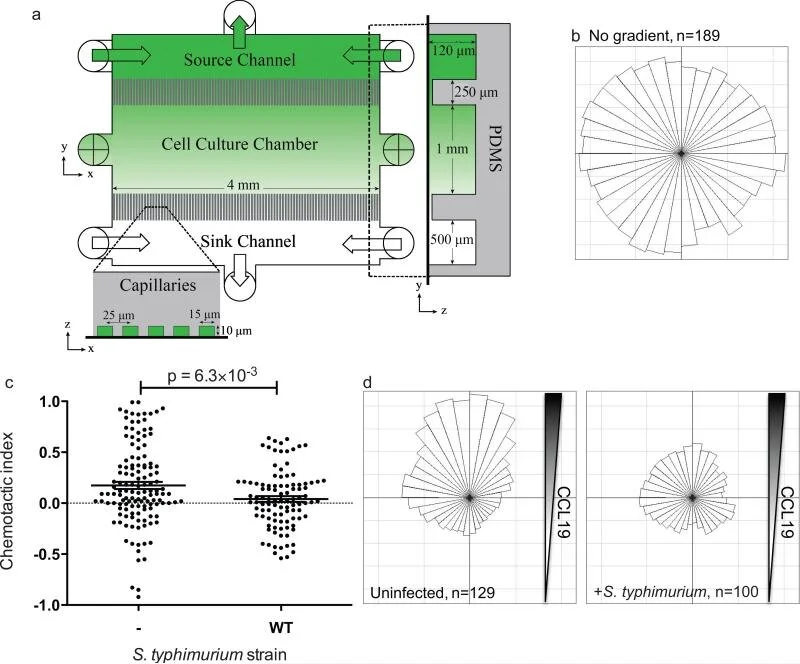
A microfluidic-based genetic screen to identify microbial virulence factors that inhibit dendritic cell migration
Microbial pathogens are able to modulate host cells and evade the immune system by multiple mechanisms. For example, Salmonella injects effector proteins into host cells and evades the host immune system in part by inhibiting dendritic cell (DC) migration. The identification of microbial factors that modulate normal host functions should lead to the development of new classes of therapeutics that target these pathways. Current screening methods to identify either host or pathogen genes involved in modulating migration towards a chemical signal are limited because they do not employ stable, precisely controlled chemical gradients. Here, we develop a positive selection microfluidic-based genetic screen that allows us to identify Salmonella virulence factors that manipulate DC migration within stable, linear chemokine gradients. Our screen identified 7 Salmonella effectors (SseF, SifA, SspH2, SlrP, PipB2, SpiC and SseI) that inhibit DC chemotaxis toward CCL19. This method is widely applicable for identifying novel microbial factors that influence normal host cell chemotaxis as well as revealing new mammalian genes involved in directed cell migration.
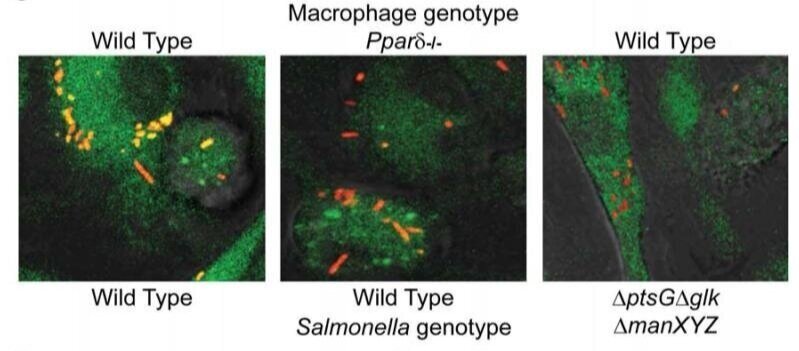
Salmonella require the fatty acid regulator PPARδ for the establishment of a metabolic environment essential for long-term persistence
Host-adapted Salmonella strains are responsible for a number of disease manifestations in mammals, including an asymptomatic chronic infection in which bacteria survive within macrophages located in systemic sites. However, the host cell physiology and metabolic requirements supporting bacterial persistence are poorly understood. In a mouse model of long-term infection, we found that S. typhimurium preferentially associates with anti-inflammatory/M2 macrophages at later stages of infection. Further, PPARδ, a eukaryotic transcription factor involved in sustaining fatty acid metabolism, is upregulated in Salmonella-infected macrophages. PPARδ deficiency dramatically inhibits Salmonella replication, which is linked to the metabolic state of macrophages and the level of intracellular glucose available to bacteria. Pharmacological activation of PPARδ increases glucose availability and enhances bacterial replication in macrophages and mice, while Salmonella fail to persist in Pparδ null mice. These data suggest that M2 macrophages represent a unique niche for long-term intracellular bacterial survival and link the PPARδ-regulated metabolic state of the host cell to persistent bacterial infection.
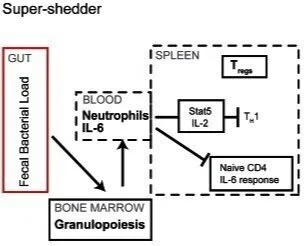
The systemic immune state of super-shedder mice is characterized by a unique neutrophil-dependent blunting of TH1 responses
Host-to-host transmission of a pathogen ensures its successful propagation and maintenance within a host population. A striking feature of disease transmission is the heterogeneity in host infectiousness. It has been proposed that within a host population, 20% of the infected hosts, termed super-shedders, are responsible for 80% of disease transmission. However, very little is known about the immune state of these super-shedders. In this study, we used the model organism Salmonella enterica serovar Typhimurium, an important cause of disease in humans and animal hosts, to study the immune state of super-shedders. Compared to moderate shedders, super-shedder mice had an active inflammatory response in both the gastrointestinal tract and the spleen but a dampened T(H)1 response specific to the secondary lymphoid organs. Spleens from super-shedder mice had higher numbers of neutrophils, and a dampened T cell response, characterized by higher levels of regulatory T cells (T(regs)), fewer T-bet(+) (T(H)1) T cells as well as blunted cytokine responsiveness. Administration of the cytokine granulocyte colony stimulating factor (G-CSF) and subsequent neutrophilia was sufficient to induce the super-shedder immune phenotype in moderate-shedder mice. Similar to super-shedders, these G-CSF-treated moderate-shedders had a dampened T(H)1 response with fewer T-bet(+) T cells and a loss of cytokine responsiveness. Additionally, G-CSF treatment inhibited IL-2-mediated TH1 expansion. Finally, depletion of neutrophils led to an increase in the number of T-bet(+) T(H)1 cells and restored their ability to respond to IL-2. Taken together, we demonstrate a novel role for neutrophils in blunting IL-2-mediated proliferation of the TH1 immune response in the spleens of mice that are colonized by high levels of S. Typhimurium in the gastrointestinal tract.
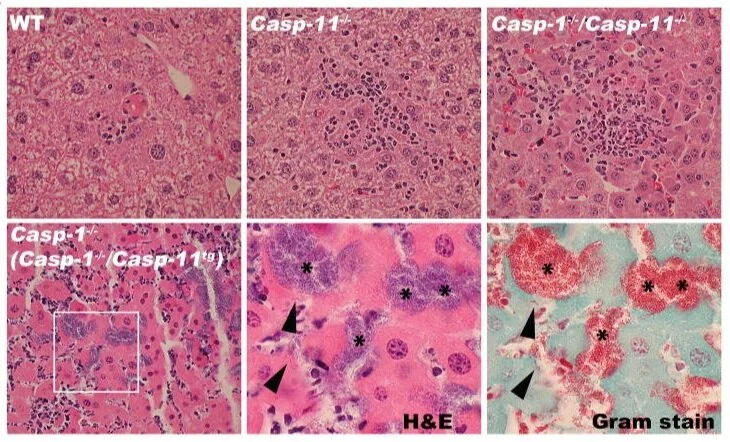
Caspase-11 increases susceptibility to Salmonella infection in the absence of caspase-1
Inflammasomes are cytosolic multiprotein complexes assembled by intracellular nucleotide-binding oligomerization domain (NOD)-like receptors (NLRs) and they initiate innate immune responses to invading pathogens and danger signals by activating caspase-1 (ref. 1). Caspase-1 activation leads to the maturation and release of the pro-inflammatory cytokines interleukin (IL)-1β and IL-18, as well as lytic inflammatory cell death known as pyroptosis. Recently, a new non-canonical inflammasome was described that activates caspase-11, a pro-inflammatory caspase required for lipopolysaccharide-induced lethality. This study also highlighted that previously generated caspase-1 knockout mice lack a functional allele of Casp11 (also known as Casp4), making them functionally Casp1 Casp11 double knockouts. Previous studies have shown that these mice are more susceptible to infections with microbial pathogens, including the bacterial pathogen Salmonella enterica serovar Typhimurium (S. typhimurium), but the individual contributions of caspase-1 and caspase-11 to this phenotype are not known. Here we show that non-canonical caspase-11 activation contributes to macrophage death during S. typhimurium infection. Toll-like receptor 4 (TLR4)-dependent and TIR-domain-containing adaptor-inducing interferon-β (TRIF)-dependent interferon-β production is crucial for caspase-11 activation in macrophages, but is only partially required for pro-caspase-11 expression, consistent with the existence of an interferon-inducible activator of caspase-11. Furthermore, Casp1(-/-) mice were significantly more susceptible to infection with S. typhimurium than mice lacking both pro-inflammatory caspases (Casp1(-/-) Casp11(-/-)). This phenotype was accompanied by higher bacterial counts, the formation of extracellular bacterial microcolonies in the infected tissue and a defect in neutrophil-mediated clearance. These results indicate that caspase-11-dependent cell death is detrimental to the host in the absence of caspase-1-mediated innate immunity, resulting in extracellular replication of a facultative intracellular bacterial pathogen.
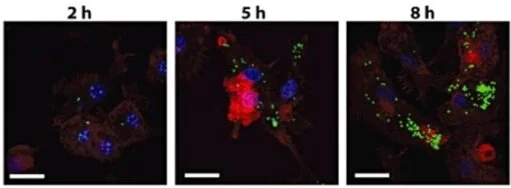
Francisella infection triggers activation of the AIM2 inflammasome in murine dendritic cells
The intracellular bacterium Francisella tularensis is the causative agent of tularemia, a potentially fatal disease. In macrophages, Francisella escapes the initial phagosome and replicates in the cytosol, where it is detected by the cytosolic DNA sensor AIM2 leading to activation of the AIM2 inflammasome. However, during aerosol infection, Francisella is also taken up by dendritic cells. In this study, we show that Francisella novicida escapes into the cytosol of bone marrow-derived dendritic cells (BMDC) where it undergoes rapid replication. We show that F. novicida activates the AIM2 inflammasome in BMDC, causing release of large amounts of IL-1β and rapid host cell death. The Francisella Pathogenicity Island is required for bacterial escape and replication and for inflammasome activation in dendritic cells. In addition, we show that bacterial DNA is bound by AIM2, which leads to inflammasome assembly in infected dendritic cells. IFN-β is upregulated in BMDC following Francisella infection, and the IFN-β signalling pathway is partially required for inflammasome activation in this cell type. Taken together, our results demonstrate that F. novicida induces inflammasome activation in dendritic cells. The resulting inflammatory cell death may be beneficial to remove the bacterial replicative niche and protect the host.
Elevated AIM2-mediated pyroptosis triggered by hypercytotoxic Francisella mutant strains is attributed to increased intracellular bacteriolysis
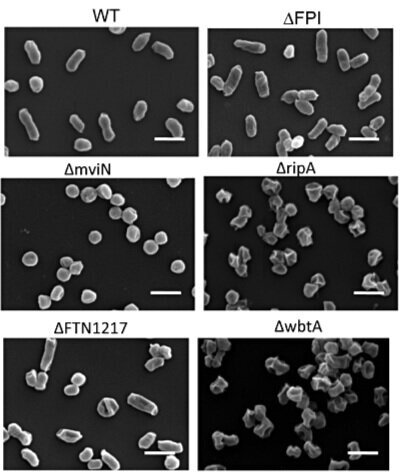
Intracellular bacterial pathogens Francisella novicida and the Live Vaccine Strain (LVS) are recognized in the macrophage cytosol by the AIM2 inflammasome, which leads to the activation of caspase-1 and the processing and secretion of active IL-1β, IL-18 and pyroptosis. Previous studies have reported that F. novicida and LVS mutants in specific genes (e.g. FTT0584, mviN and ripA) induce elevated inflammasome activation and hypercytotoxicity in host cells, leading to the proposal that F. novicida and LVS may have proteins that actively modulate inflammasome activation. However, there has been no direct evidence of such inflammasome evasion mechanisms. Here, we demonstrate for the first time that the above mutants, along with a wide range of F. novicida hypercytotoxic mutants that are deficient for membrane-associated proteins (ΔFTT0584, ΔmviN, ΔripA, ΔfopA and ΔFTN1217) or deficient for genes involved in O-antigen or LPS biosynthesis (ΔwbtA and ΔlpxH) lyse more intracellularly, thus activating increased levels of AIM2-dependent pyroptosis and other innate immune signalling pathways. This suggests that an inflammasome-specific evasion mechanism may not be present in F. novicida and LVS. Furthermore, future studies may need to consider increased bacterial lysis as a possible cause of elevated stimulation of multiple innate immune pathways when the protein composition or surface carbohydrates of the bacterial membrane is altered.
Differential Requirement for Caspase-1 Autoproteolysis in Pathogen-Induced Cell Death and Cytokine Processing
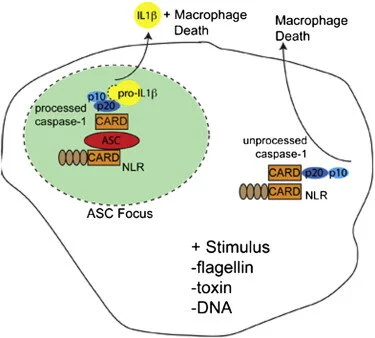
Activation of the cysteine protease Caspase-1 is a key event in the innate immune response to infections. Synthesized as a proprotein, Caspase-1 undergoes autoproteolysis within multiprotein complexes called inflammasomes. Activated Caspase-1 is required for proteolytic processing and for release of the cytokines interleukin-1β and interleukin-18, and it can also cause rapid macrophage cell death. We show that macrophage cell death and cytokine maturation in response to infection with diverse bacterial pathogens can be separated genetically and that two distinct inflammasome complexes mediate these events. Inflammasomes containing the signaling adaptor Asc form a single large "focus" in which Caspase-1 undergoes autoproteolysis and processes IL-1β/IL-18. In contrast, Asc-independent inflammasomes activate Caspase-1 without autoproteolysis and do not form any large structures in the cytosol. Caspase-1 mutants unable to undergo autoproteolysis promoted rapid cell death, but processed IL-1β/18 inefficiently. Our results suggest the formation of spatially and functionally distinct inflammasomes complexes in response to bacterial pathogens.
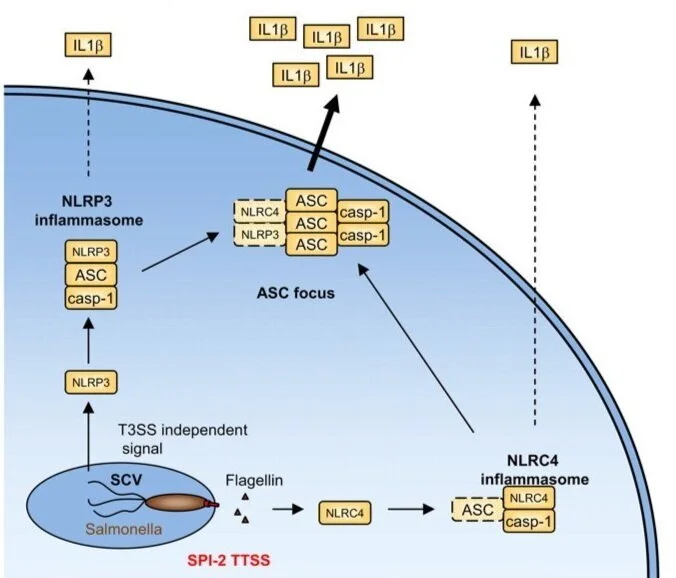
Redundant roles for inflammasome receptors NLRP3 and NLRC4 in host defense against Salmonella
Journal of Experimental Medicine, 2010
Intracellular pathogens and endogenous danger signals in the cytosol engage NOD-like receptors (NLRs), which assemble inflammasome complexes to activate caspase-1 and promote the release of proinflammatory cytokines IL-1β and IL-18. However, the NLRs that respond to microbial pathogens in vivo are poorly defined. We show that the NLRs NLRP3 and NLRC4 both activate caspase-1 in response to Salmonella typhimurium. Responding to distinct bacterial triggers, NLRP3 and NLRC4 recruited ASC and caspase-1 into a single cytoplasmic focus, which served as the site of pro-IL-1β processing. Consistent with an important role for both NLRP3 and NLRC4 in innate immune defense against S. typhimurium, mice lacking both NLRs were markedly more susceptible to infection. These results reveal unexpected redundancy among NLRs in host defense against intracellular pathogens in vivo.
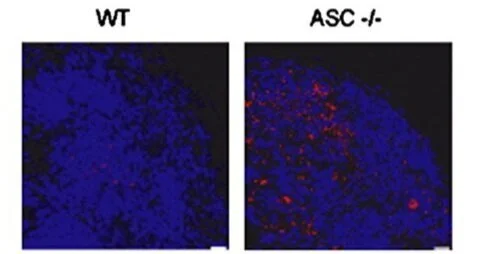
Absent in melanoma 2 is required for innate immune recognition of Francisella tularensis
Proceedings of the National Academy of Sciences, 2010
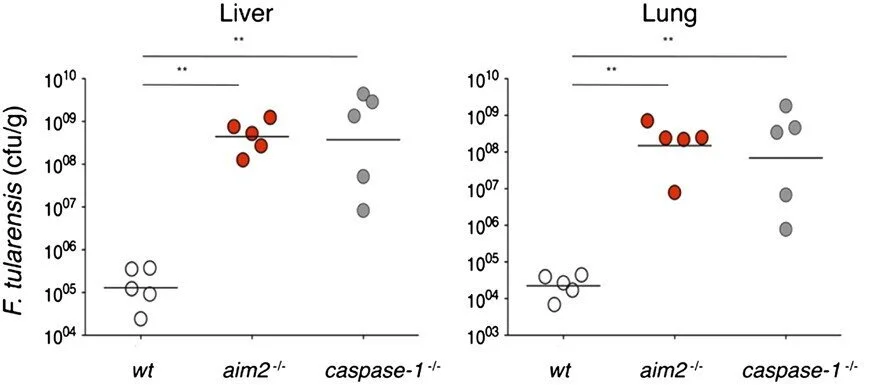
Macrophages respond to cytosolic nucleic acids by activating cysteine protease caspase-1 within a complex called the inflammasome. Subsequent cleavage and secretion of proinflammatory cytokines IL-1β and IL-18 are critical for innate immunity. Here, we show that macrophages from mice lacking absent in melanoma 2 (AIM2) cannot sense cytosolic double-stranded DNA and fail to trigger inflammasome assembly. Caspase-1 activation in response to intracellular pathogen Francisella tularensis also required AIM2. Immunofluorescence microscopy of macrophages infected with F. tularensis revealed striking colocalization of bacterial DNA with endogenous AIM2 and inflammasome adaptor ASC. By contrast, type I IFN (IFN-α and -β) secretion in response to F. tularensis did not require AIM2. IFN-I did, however, boost AIM2-dependent caspase-1 activation by increasing AIM2 protein levels. Thus, inflammasome activation was reduced in infected macrophages lacking either the IFN-I receptor or stimulator of interferon genes (STING). Finally, AIM2-deficient mice displayed increased susceptibility to F. tularensis infection compared with wild-type mice. Their increased bacterial burden in vivo confirmed that AIM2 is essential for an effective innate immune response.
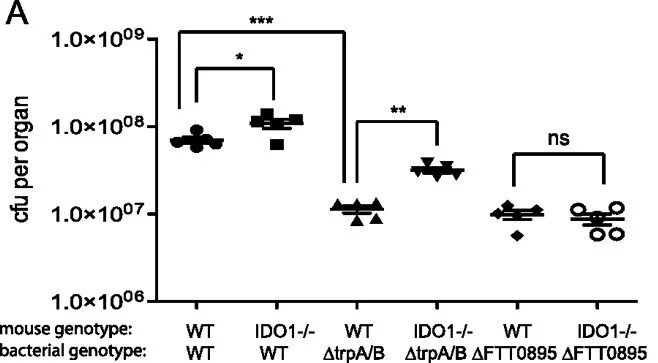
Indoleamine 2,3-Dioxygenase 1 Is a Lung-Specific Innate Immune Defense Mechanism That Inhibits Growth of Francisella tularensis Tryptophan Auxotrophs
Upon microbial challenge, organs at various anatomic sites of the body employ different innate immune mechanisms to defend against potential infections. Accordingly, microbial pathogens evolved to subvert these organ-specific host immune mechanisms to survive and grow in infected organs. Francisella tularensis is a bacterium capable of infecting multiple organs and thus encounters a myriad of organ-specific defense mechanisms. This suggests that F. tularensis may possess specific factors that aid in evasion of these innate immune defenses. We carried out a microarray-based, negative-selection screen in an intranasal model of Francisella novicida infection to identify Francisella genes that contribute to bacterial growth specifically in the lungs of mice. Genes in the bacterial tryptophan biosynthetic pathway were identified as being important for F. novicida growth specifically in the lungs. In addition, a host tryptophan-catabolizing enzyme, indoleamine 2,3-dioxygenase 1 (IDO1), is induced specifically in the lungs of mice infected with F. novicida or Streptococcus pneumoniae. Furthermore, the attenuation of F. novicida tryptophan mutant bacteria was rescued in the lungs of IDO1−/− mice. IDO1 is a lung-specific innate immune mechanism that controls pulmonary Francisella infections.
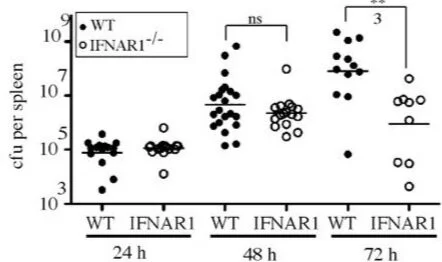
Type I IFN Signaling Constrains IL-17A/F Secretion by γδ T Cells during Bacterial Infections
Recognition of intracellular bacteria by macrophages leads to secretion of type I IFNs. However, the role of type I IFN during bacterial infection is still poorly understood. Francisella tularensis, the causative agent of tularemia, is a pathogenic bacterium that replicates in the cytosol of macrophages leading to secretion of type I IFN. In this study, we investigated the role of type I IFNs in a mouse model of tularemia. Mice deficient for type I IFN receptor (IFNAR1−/−) are more resistant to intradermal infection with F. tularensis subspecies novicida (F. novicida). Increased resistance to infection was associated with a specific increase in IL-17A/F and a corresponding expansion of an IL-17A+ γδ T cell population, indicating that type I IFNs negatively regulate the number of IL-17A+ γδ T cells during infection. Furthermore, IL-17A–deficient mice contained fewer neutrophils compared with wild-type mice during infection, indicating that IL-17A contributes to neutrophil expansion during F. novicida infection. Accordingly, an increase in IL-17A in IFNAR1−/− mice correlated with an increase in splenic neutrophil numbers. Similar results were obtained in a mouse model of pneumonic tularemia using the highly virulent F. tularensis subspecies tularensis SchuS4 strain and in a mouse model of systemic Listeria monocytogenes infection. Our results indicate that the type I IFN-mediated negative regulation of IL-17A+ γδ T cell expansion is conserved during bacterial infections. We propose that this newly described activity of type I IFN signaling might participate in the resistance of the IFNAR1−/− mice to infection with F. novicida and other intracellular bacteria.
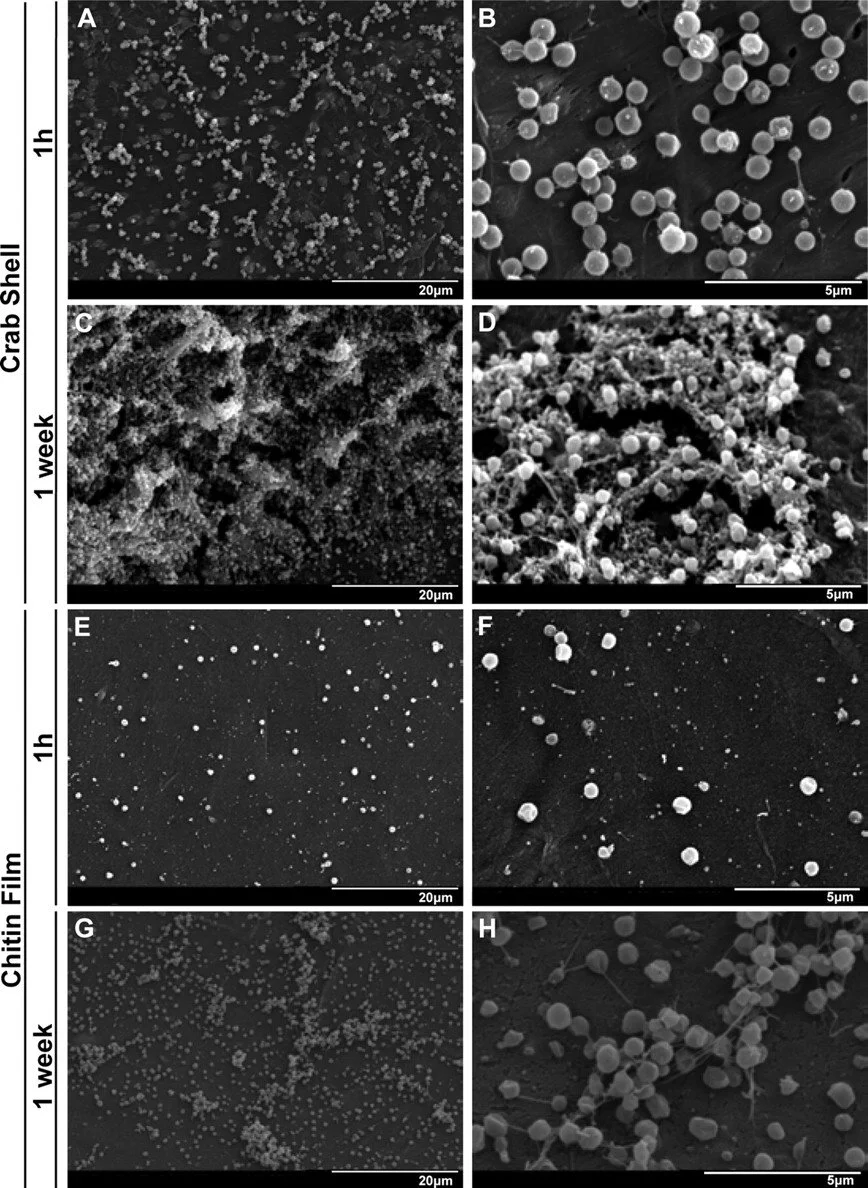
Contributions of Francisella tularensis subsp. Novicida Chitinases and Sec Secretion System to Biofilm Formation on Chitin
Applied and Environmental Microbiology, 2010
Francisella tularensis, the zoonotic cause of tularemia, can infect numerous mammals and other eukaryotes. Although studying F. tularensis pathogenesis is essential to comprehending disease, mammalian infection is just one step in the ecology of Francisella species. F. tularensis has been isolated from aquatic environments and arthropod vectors, environments in which chitin could serve as a potential carbon source and as a surface for attachment and growth. We show that F. tularensis subsp. novicida forms biofilms during the colonization of chitin surfaces. The ability of F. tularensis to persist using chitin as a sole carbon source is dependent on chitinases, since mutants lacking chiA or chiB are attenuated for chitin colonization and biofilm formation in the absence of exogenous sugar. A genetic screen for biofilm mutants identified the Sec translocon export pathway and 14 secreted proteins. We show that these genes are important for initial attachment during biofilm formation. We generated defined deletion mutants by targeting two chaperone genes (secB1 and secB2) involved in Sec-dependent secretion and four genes that encode putative secreted proteins. All of the mutants were deficient in attachment to polystyrene and chitin surfaces and for biofilm formation compared to wild-type F. novicida. In contrast, mutations in the Sec translocon and secreted factors did not affect virulence. Our data suggest that biofilm formation by F. tularensis promotes persistence on chitin surfaces. Further study of the interaction of F. tularensis with the chitin microenvironment may provide insight into the environmental survival and transmission mechanisms of this pathogen.
The Salmonella SPI2 Effector SseI Mediates Long-Term Systemic Infection by Modulating Host Cell Migration
Host-adapted strains of Salmonella enterica cause systemic infections and have the ability to persist systemically for long periods of time despite the presence of a robust immune response. Chronically infected hosts are asymptomatic and transmit disease to naïve hosts via fecal shedding of bacteria, thereby serving as a critical reservoir for disease. We show that the bacterial effector protein SseI (also called SrfH), which is translocated into host cells by the Salmonella Pathogenicity Island 2 (SPI2) type III secretion system (T3SS), is required for Salmonella typhimurium to maintain a long-term chronic systemic infection in mice. SseI inhibits normal cell migration of primary macrophages and dendritic cells (DC) in vitro, and such inhibition requires the host factor IQ motif containing GTPase activating protein 1 (IQGAP1), an important regulator of cell migration. SseI binds directly to IQGAP1 and co-localizes with this factor at the cell periphery. The C-terminal domain of SseI is similar to PMT/ToxA, a bacterial toxin that contains a cysteine residue (C1165) that is critical for activity. Mutation of the corresponding residue in SseI (C178A) eliminates SseI function in vitro and in vivo, but not binding to IQGAP1. In addition, infection with wild-type (WT) S. typhimurium suppressed DC migration to the spleen in vivo in an SseI-dependent manner. Correspondingly, examination of spleens from mice infected with WT S. typhimurium revealed fewer DC and CD4+ T lymphocytes compared to mice infected with ΔsseI S. typhimurium. Taken together, our results demonstrate that SseI inhibits normal host cell migration, which ultimately counteracts the ability of the host to clear systemic bacteria.
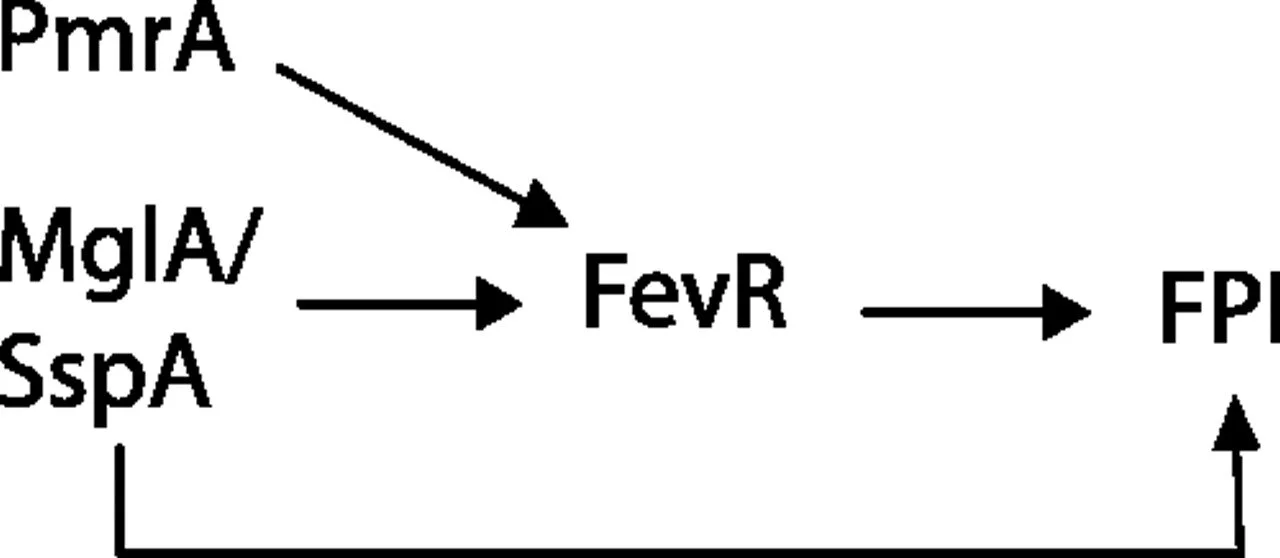
Identification of fevR, a Novel Regulator of Virulence Gene Expression in Francisella novicida
Francisella tularensis infects wild animals and humans to cause tularemia. This pathogen targets the cytosol of macrophages, where it replicates using the genes in the Francisella pathogenicity island (FPI). Virulence gene regulation in Francisella is complex, but transcriptional regulators MglA and SspA have been shown to regulate the expression of approximately 100 genes, including the entire FPI. We utilized a Francisella novicida transposon mutant library to identify additional regulatory factors and identified five additional genes that are essential for virulence gene expression. One regulatory gene, FTN_0480 (fevR, Francisella effector of virulence regulation), present in all Francisella species, is required for expression of the FPI genes and other genes in the MglA/SspA regulon. The expression of fevR is positively regulated by MglA. However, constitutive expression of fevR in an mglA mutant strain did not restore expression of the MglA/SspA regulon, demonstrating that mglA and fevR act in parallel to positively regulate virulence gene expression. Virulence studies revealed that fevR is essential for bacterial replication in macrophages and in mice, where we additionally show that fevR is required for the expression of genes in the MglA/SspA regulon in vivo. Thus, fevR is a crucial virulence gene in Francisella, required for the expression of virulence factors known to be essential for this pathogen's subversion of host defenses and pathogenesis in vivo.
Host Transmission of Salmonella enterica Serovar Typhimurium Is Controlled by Virulence Factors and Indigenous Intestinal Microbiota
Transmission is an essential stage of a pathogen's life cycle and remains poorly understood. We describe here a model in which persistently infected 129X1/SvJ mice provide a natural model of Salmonella enterica serovar Typhimurium transmission. In this model only a subset of the infected mice, termed supershedders, shed high levels (>108 CFU/g) of Salmonella serovar Typhimurium in their feces and, as a result, rapidly transmit infection. While most Salmonella serovar Typhimurium-infected mice show signs of intestinal inflammation, only supershedder mice develop colitis. Development of the supershedder phenotype depends on the virulence determinants Salmonella pathogenicity islands 1 and 2, and it is characterized by mucosal invasion and, importantly, high luminal abundance of Salmonella serovar Typhimurium within the colon. Immunosuppression of infected mice does not induce the supershedder phenotype, demonstrating that the immune response is not the main determinant of Salmonella serovar Typhimurium levels within the colon. In contrast, treatment of mice with antibiotics that alter the health-associated indigenous intestinal microbiota rapidly induces the supershedder phenotype in infected mice and predisposes uninfected mice to the supershedder phenotype for several days. These results demonstrate that the intestinal microbiota plays a critical role in controlling Salmonella serovar Typhimurium infection, disease, and transmissibility. This novel model should facilitate the study of host, pathogen, and intestinal microbiota factors that contribute to infectious disease transmission.
Type I interferon signaling is required for activation of the inflammasome during Francisella infection
Journal of Experimental Medicine, 2007
Francisella tularensis is a pathogenic bacterium whose virulence is linked to its ability to replicate within the host cell cytosol. Entry into the macrophage cytosol activates a host-protective multimolecular complex called the inflammasome to release the proinflammatory cytokines interleukin (IL)-1β and -18 and trigger caspase-1–dependent cell death. In this study, we show that cytosolic F. tularensis subspecies novicida (F. novicida) induces a type I interferon (IFN) response that is essential for caspase-1 activation, inflammasome-mediated cell death, and release of IL-1β and -18. Extensive type I IFN–dependent cell death resulting in macrophage depletion occurs in vivo during F. novicida infection. Type I IFN is also necessary for inflammasome activation in response to cytosolic Listeria monocytogenes but not vacuole-localized Salmonella enterica serovar Typhimurium or extracellular adenosine triphosphate. These results show the specific connection between type I IFN signaling and inflammasome activation, which are two sequential events triggered by the recognition of cytosolic bacteria. To our knowledge, this is the first example of the positive regulation of inflammasome activation. This connection underscores the importance of the cytosolic recognition of pathogens and highlights how multiple innate immunity pathways interact before commitment to critical host responses.
In vivo negative selection screen identifies genes required for Francisella virulence
Proceedings of the National Academy of Sciences, 2007
Francisella tularensis subverts the immune system to rapidly grow within mammalian hosts, often causing tularemia, a fatal disease. This pathogen targets the cytosol of macrophages where it replicates by using the genes encoded in the Francisella pathogenicity island. However, the bacteria are recognized in the cytosol by the host's ASC/caspase-1 pathway, which is essential for host defense, and leads to macrophage cell death and proinflammatory cytokine production. We used a microarray-based negative selection screen to identify Francisella genes that contribute to growth and/or survival in mice. The screen identified many known virulence factors including all of the Francisella pathogenicity island genes, LPS O-antigen synthetic genes, and capsule synthetic genes. We also identified 44 previously unidentified genes that were required for Francisella virulence in vivo, indicating that this pathogen may use uncharacterized mechanisms to cause disease. Among these, we discovered a class of Francisella virulence genes that are essential for growth and survival in vivo but do not play a role in intracellular replication within macrophages. Instead, these genes modulate the host ASC/caspase-1 pathway, a previously unidentified mechanism of Francisella pathogenesis. This finding indicates that the elucidation of the molecular mechanisms used by other uncharacterized genes identified in our screen will increase our understanding of the ways in which bacterial pathogens subvert the immune system.
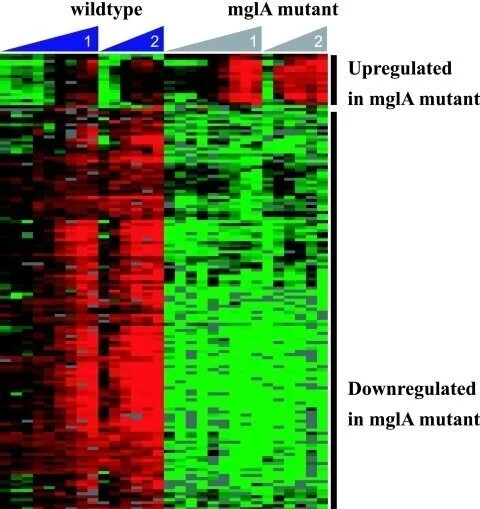
Identification of MglA-Regulated Genes Reveals Novel Virulence Factors in Francisella tularensis
The facultative intracellular bacterium Francisella tularensis causes the zoonotic disease tularemia. F. tularensis resides within host macrophages in vivo, and this ability is essential for pathogenesis. The transcription factor MglA is required for the expression of several Francisella genes that are necessary for replication in macrophages and for virulence in mice. We hypothesized that the identification of MglA-regulated genes in the Francisella genome by transcriptional profiling of wild-type and mglA mutant bacteria would lead to the discovery of new virulence factors utilized by F. tularensis. A total of 102 MglA-regulated genes were identified, the majority of which were positively regulated, including all of the Francisella pathogenicity island (FPI) genes. We mutated novel MglA-regulated genes and tested the mutants for their ability to replicate and induce cytotoxicity in macrophages and to grow in mice. Mutations in MglA-regulated genes within the FPI (pdpB and cds2) as well as outside the FPI (FTT0989, oppB, and FTT1209c) were either attenuated or hypervirulent in macrophages compared to the wild-type strain. All of these mutants exhibited decreased fitness in vivo in competition experiments with wild-type bacteria. We have identified five new Francisella virulence genes, and our results suggest that characterizations of additional MglA-regulated genes will yield further insights into the pathogenesis of this bacterium.
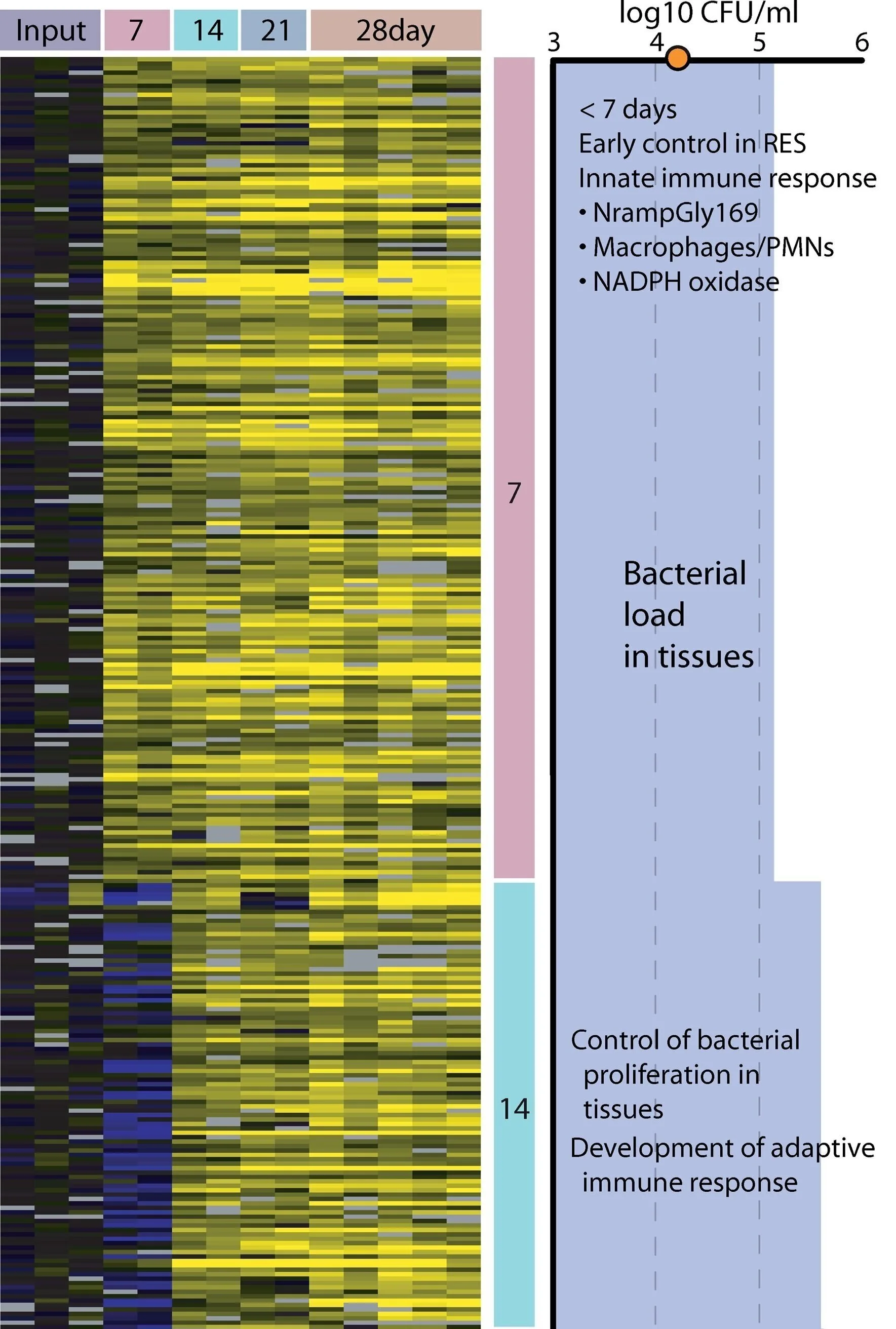
Genome-Wide Screen for Salmonella Genes Required for Long-Term Systemic Infection of the Mouse
A microarray-based negative selection screen was performed to identify Salmonella enterica serovar Typhimurium (serovar Typhimurium) genes that contribute to long-term systemic infection in 129X1/SvJ (Nramp1r) mice. A high-complexity transposon-mutagenized library was used to infect mice intraperitoneally, and the selective disappearance of mutants was monitored after 7, 14, 21, and 28 d postinfection. One hundred and eighteen genes were identified to contribute to serovar Typhimurium infection of the spleens of mice by 28 d postinfection. The negatively selected mutants represent many known aspects of Salmonella physiology and pathogenesis, although the majority of the identified genes are of putative or unknown function. Approximately 30% of the negatively selected genes correspond to horizontally acquired regions such as those within Salmonella pathogenicity islands (SPI 1-5), prophages (Gifsy-1 and -2 and remnant), and the pSLT virulence plasmid. In addition, mutations in genes responsible for outer membrane structure and remodeling, such as LPS- and PhoP-regulated and fimbrial genes, were also selected against. Competitive index experiments demonstrated that the secreted SPI2 effectors SseK2 and SseJ as well as the SPI4 locus are attenuated relative to wild-type bacteria during systemic infection. Interestingly, several SPI1-encoded type III secretion system effectors/translocases are required by serovar Typhimurium to establish and, unexpectedly, to persist systemically, challenging the present description of Salmonella pathogenesis. Moreover, we observed a progressive selection against serovar Typhimurium mutants based upon the duration of the infection, suggesting that different classes of genes may be required at distinct stages of infection. Overall, these data indicate that Salmonella long-term systemic infection in the mouse requires a diverse repertoire of virulence factors. This diversity of genes presumably reflects the fact that bacteria sequentially encounter a variety of host environments and that Salmonella has evolved to respond to these selective forces in a way that permits both the bacteria and the host to survive.
Innate immunity against Francisella tularensis is dependent on the ASC/caspase-1 axis
Journal of Experimental Medicine, 2005
Francisella tularensis is a highly infectious gram-negative coccobacillus that causes the zoonosis tularemia. This bacterial pathogen causes a plague-like disease in humans after exposure to as few as 10 cells. Many of the mechanisms by which the innate immune system fights Francisella are unknown. Here we show that wild-type Francisella, which reach the cytosol, but not Francisella mutants that remain localized to the vacuole, induced a host defense response in macrophages, which is dependent on caspase-1 and the death-fold containing adaptor protein ASC. Caspase-1 and ASC signaling resulted in host cell death and the release of the proinflammatory cytokines interleukin (IL)-1β and IL-18. F. tularensis–infected caspase-1– and ASC-deficient mice showed markedly increased bacterial burdens and mortality as compared with wild-type mice, demonstrating a key role for caspase-1 and ASC in innate defense against infection by this pathogen.
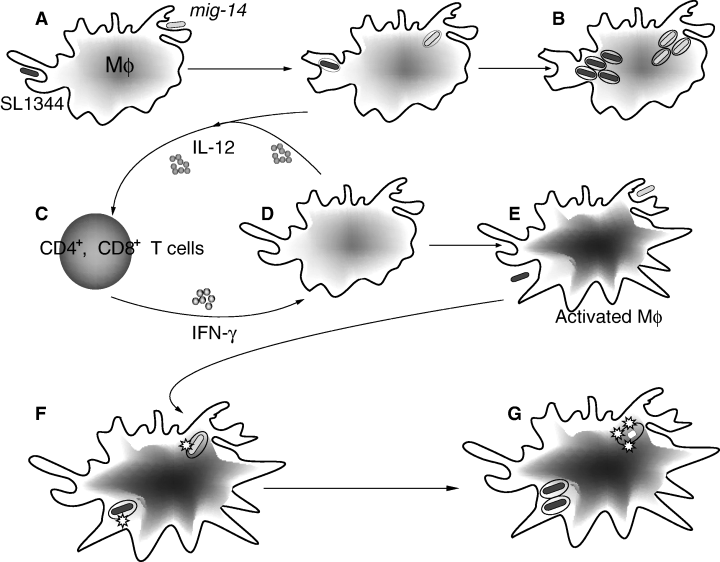
Mig-14 is an inner membrane-associated protein that promotes Salmonella typhimurium resistance to CRAMP, survival within activated macrophages and persistent infection
Salmonella enterica serovar Typhimurium (S. typhimurium) infects a wide variety of mammalian hosts and in rodents causes a typhoid-like systemic disease involving replication of bacteria inside macrophages within reticuloendothelial tissues. Previous studies demonstrated that the mig-14 and virK genes of Salmonella enterica are important in bacterial resistance to anti-microbial peptides and are necessary for continued replication of S. typhimurium in the liver and spleen of susceptible mice after orogastric inoculation. In this work we report that inflammatory signalling via interferon-gamma (IFN-γ) is crucial to controlling replication of mig-14 mutant bacteria within the liver and spleen of mice after oral infection. Using a Salmonella persistence model recently developed in our laboratory, we further demonstrate that mig-14 contributes to long-term persistence of Salmonella in the spleen and mesenteric lymph nodes of chronically infected mice. Both mig-14 and virK contribute to the survival of Salmonella in macrophages treated with IFN-γ and are necessary for resistance to cathelin-related anti-microbial peptide (CRAMP), an anti-microbial peptide expressed at high levels in activated mouse macrophages. We also show that both Mig-14 and VirK inhibit the binding of CRAMP to Salmonella, and demonstrate that Mig-14 is an inner membrane-associated protein. We further demonstrate by transmission electron microscopy that the primary locus of CRAMP activity appears to be intracytoplasmic, rather than at the outer membrane, suggesting that Mig-14 may prevent the penetration of the inner membrane by CRAMP. Together, these data indicate an important role for mig-14 in anti-microbial peptide resistance in vivo, and show that this resistance is important to the survival of Salmonella in systemic sites during both acute and persistent infection.
Salmonella typhimurium Persists within Macrophages in the Mesenteric Lymph Nodes of Chronically Infected Nramp1+/+ Mice and Can Be Reactivated by IFNγ Neutralization
Journal of Experimental Medicine, 2004
Host-adapted strains of Salmonella are capable of establishing a persistent infection in their host often in the absence of clinical disease. The mouse model of Salmonella infection has primarily been used as a model for the acute systemic disease. Therefore, the sites of long-term S. typhimurium persistence in the mouse are not known nor are the mechanisms of persistent infection clearly understood. Here, we show that S. typhimurium can persist for as long as 1 yr in the mesenteric lymph nodes (MLNs) of 129sv Nramp1+/+ (Slc11a1+/+) mice despite the presence of high levels of anti–S. typhimurium antibody. Tissues from 129sv mice colonized for 60 d contain numerous inflammatory foci and lesions with features resembling S. typhi granulomas. Tissues from mice infected for 365 d have very few organized inflammatory lesions, but the bacteria continue to persist within macrophages in the MLN and the animals generally remain disease-free. Finally, chronically infected mice treated with an interferon-γ neutralizing antibody exhibited symptoms of acute systemic infection, with evidence of high levels of bacterial replication in most tissues and high levels of fecal shedding. Thus, interferon-γ, which may affect the level of macrophage activation, plays an essential role in the control of the persistent S. typhimurium infection in mice.
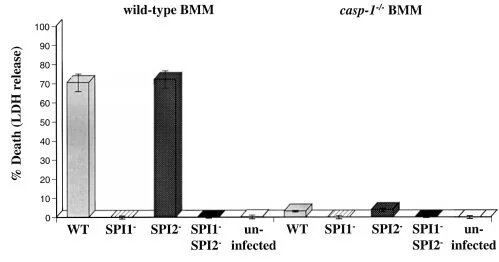
Salmonella pathogenicity island 2-dependent macrophage death is mediated in part by the host cysteine protease caspase-1
Salmonella typhimurium invades host macrophages and can either induce a rapid cell death or establish an intracellular niche within the phagocytic vacuole. Rapid cell death requires the Salmonella pathogenicity island (SPI)1 and the host protein caspase-1, a member of the pro-apoptotic caspase family of proteases. Salmonella that do not cause this rapid cell death and instead reside in the phagocytic vacuole can trigger macrophage death at a later time point. We show here that the human pathogen Salmonella typhi also triggers both rapid, caspase-1-dependent and delayed cell death in human monocytes. The delayed cell death has previously been shown with S. typhimurium to be dependent on SPI2-encoded genes and ompR. Using caspase-1–/– bone marrow-derived macrophages and isogenic S. typhimurium mutant strains, we show that a large portion of the delayed, SPI2-dependent death is mediated by caspase-1. The two known substrates of activated caspase-1 are the pro-inflammatory cytokines interleukin-1β (IL-1β) and IL-18, which are cleaved to produce bioactive cytokines. We show here that IL-1β is released during both SPI1- and SPI2-dependent macrophage killing. Using IL-1β–/– bone marrow-derived macrophages and a neutralizing anti-IL-18 antibody, we show that neither IL-1β nor IL-18 is required for rapid or delayed macrophage death. Thus, both rapid, SPI1-mediated killing and delayed, SPI2-mediated killing require caspase-1 and result in the secretion of IL-1β, which promotes inflammation and may facilitate the spread of Salmonella beyond the gastrointestinal tract in systemic disease.
Salmonella Exploits Caspase-1 to Colonize Peyer's Patches in a Murine Typhoid Model
Journal of Experimental Medicine, 2000
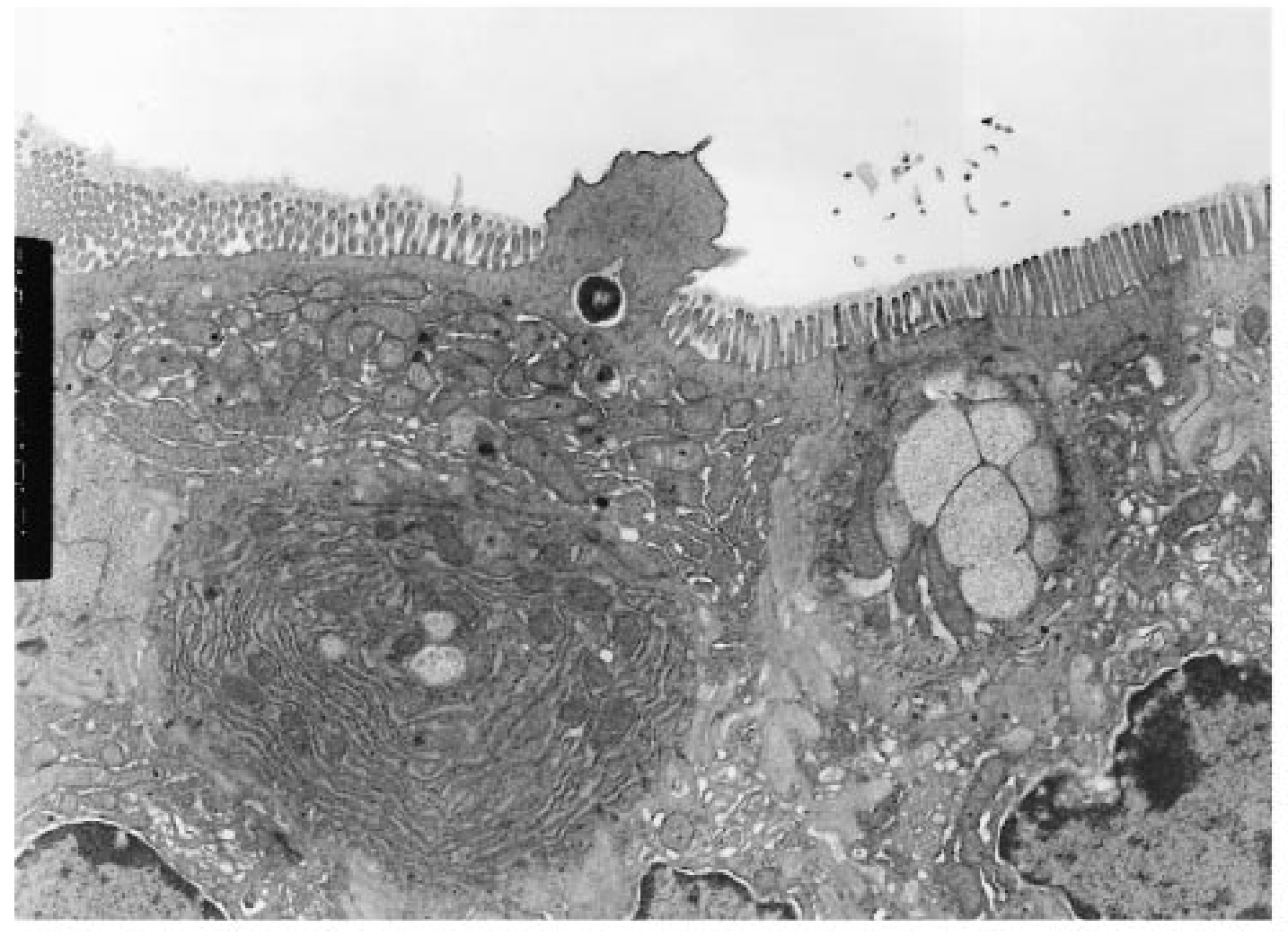
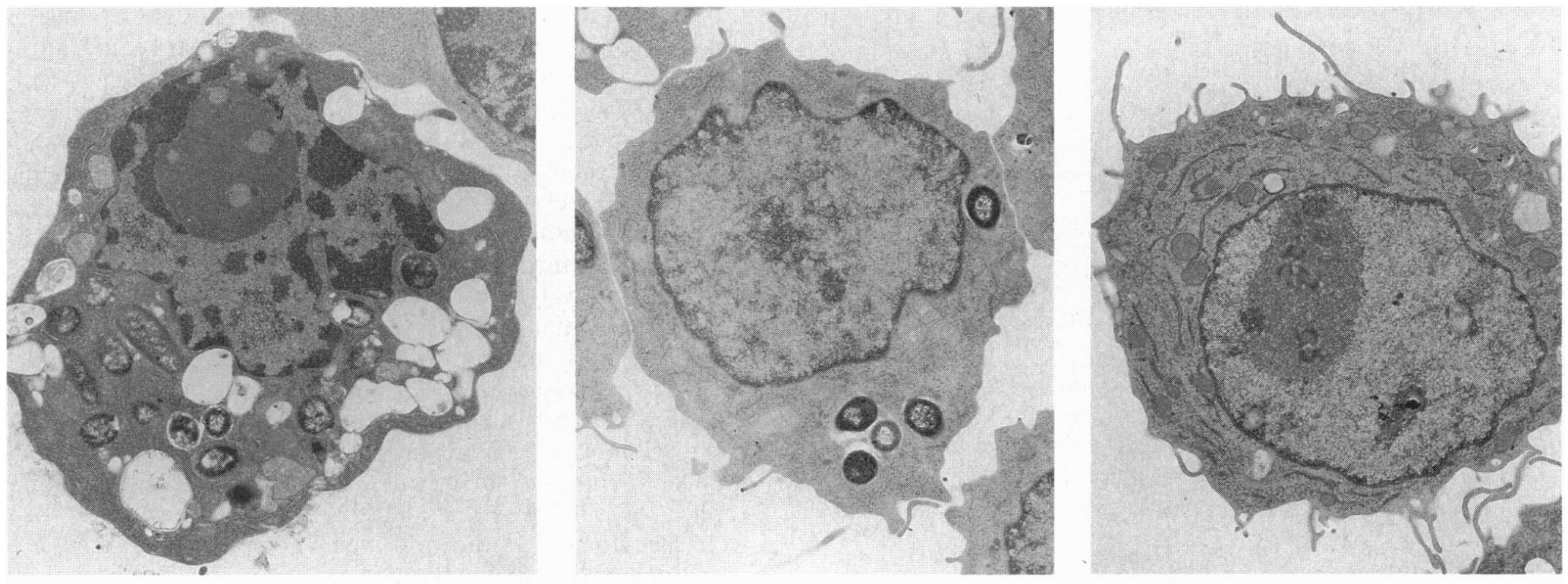
Salmonella typhimurium invades host macrophages and induces apoptosis and the release of mature proinflammatory cytokines. SipB, a protein translocated by Salmonella into the cytoplasm of macrophages, is required for activation of Caspase-1 (Casp-1, an interleukin [IL]-1β–converting enzyme), which is a member of a family of cysteine proteases that induce apoptosis in mammalian cells. Casp-1 is unique among caspases because it also directly cleaves the proinflammatory cytokines IL-1β and IL-18 to produce bioactive cytokines. We show here that mice lacking Casp-1 (casp-1−/− mice) had an oral S. typhimurium 50% lethal dose (LD50) that was 1,000-fold higher than that of wild-type mice. Salmonella breached the M cell barrier of casp-1−/− mice efficiently; however, there was a decrease in the number of apoptotic cells, intracellular bacteria, and the recruitment of polymorphonuclear lymphocytes in the Peyer's patches (PP) as compared with wild-type mice. Furthermore, Salmonella did not disseminate systemically in the majority of casp-1−/− mice, as demonstrated by significantly less colonization in the PP, mesenteric lymph nodes, and spleens of casp-1−/− mice after an oral dose of S. typhimurium that was 100-fold higher than the LD50. The increased resistance in casp-1−/− animals appears specific for Salmonella infection since these mice were susceptible to colonization by another enteric pathogen, Yersinia pseudotuberculosis, which normally invades the PP. These results show that Casp-1, which is both proapoptotic and proinflammatory, is essential for S. typhimurium to efficiently colonize the cecum and PP and subsequently cause systemic typhoid-like disease in mice.
Salmonella typhimurium invasion induces apoptosis in infected macrophages
Proceedings of the National Academy of Sciences, 1996
Invasive Salmonella typhimurium induces dramatic cytoskeletal changes on the membrane surface of mammalian epithelial cells and RAW264.7 macrophages as part of its entry mechanism. Noninvasive S. typhimurium strains are unable to induce this membrane ruffling. Invasive S. typhimurium strains invade RAW264.7 macrophages in 2 h with 7- to 10-fold higher levels than noninvasive strains. Invasive S. typhimurium and Salmonella typhi, independent of their ability to replicate intracellularly, are cytotoxic to RAW264.7 macrophages and, to a greater degree, to murine bone marrow-derived macrophages. Here, we show that the macrophage cytotoxicity mediated by invasive Salmonella is apoptosis, as shown by nuclear morphology, cytoplasmic vacuolization, and host cell DNA fragmentation. S. typhimurium that enter cells causing ruffles but are mutant for subsequent intracellular replication also initiate host cell apoptosis. Mutant S. typhimurium that are incapable of inducing host cell membrane ruffling fail to induce apoptosis. The activation state of the macrophage plays a significant role in the response of macrophages to Salmonella invasion, perhaps indicating that the signal or receptor for initiating programmed cell death is upregulated in activated macrophages. The ability of Salmonella to promote apoptosis may be important for the initiation of infection, bacterial survival, and escape of the host immune response.